Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
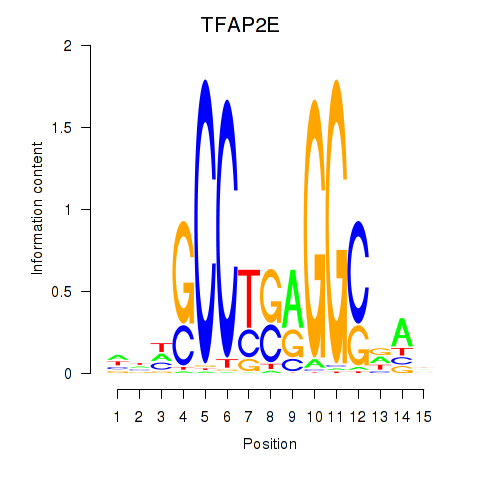
Results for TFAP2E
Z-value: 0.50
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
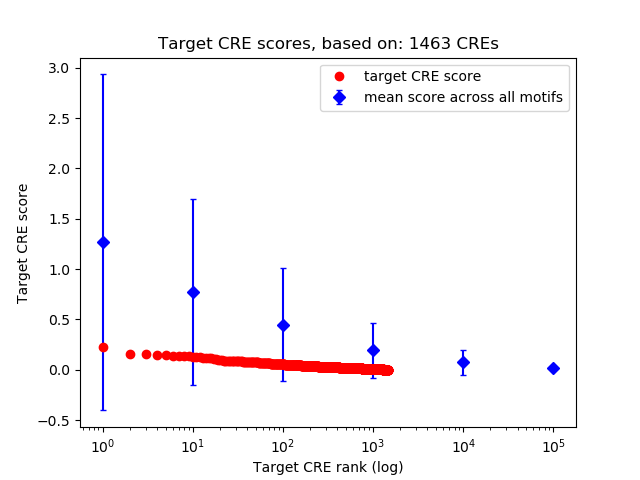
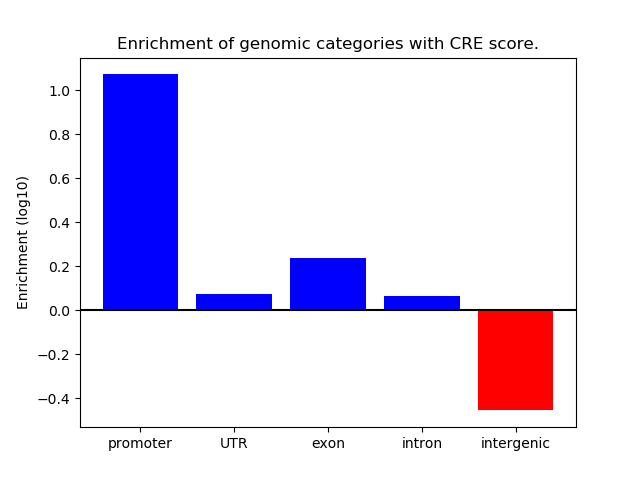
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_36038362_36038513 | TFAP2E | 534 | 0.574435 | -0.20 | 6.1e-01 | Click! |
| chr1_36038675_36039393 | TFAP2E | 63 | 0.797400 | -0.20 | 6.1e-01 | Click! |
Activity of the TFAP2E motif across conditions
Conditions sorted by the z-value of the TFAP2E motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
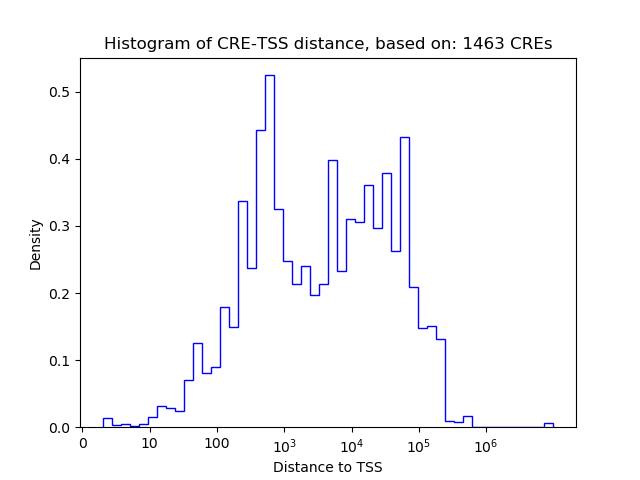
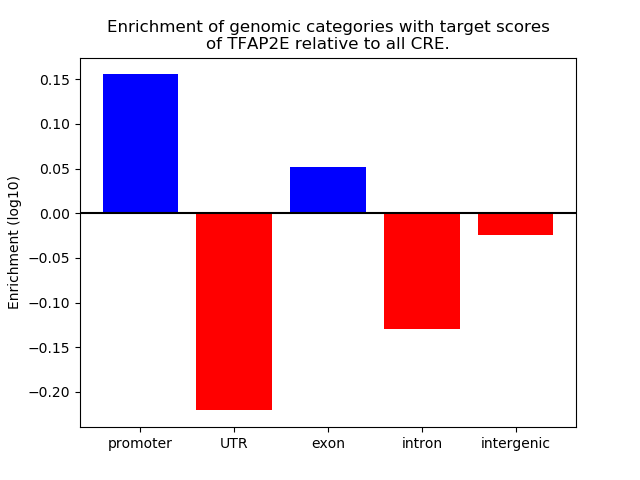
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_153406166_153406317 | 0.23 |
FMNL2 |
formin-like 2 |
69851 |
0.13 |
| chr8_57594130_57594603 | 0.16 |
RP11-17A4.2 |
|
192709 |
0.03 |
| chr9_35489468_35490886 | 0.15 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr3_192549660_192549811 | 0.14 |
FGF12 |
fibroblast growth factor 12 |
64182 |
0.13 |
| chr11_119189616_119189883 | 0.14 |
MCAM |
melanoma cell adhesion molecule |
1923 |
0.17 |
| chr5_34710476_34710647 | 0.14 |
RAI14 |
retinoic acid induced 14 |
22897 |
0.22 |
| chr1_201436884_201437966 | 0.14 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
887 |
0.55 |
| chr14_61123462_61124719 | 0.14 |
SIX1 |
SIX homeobox 1 |
887 |
0.64 |
| chr1_118644330_118644506 | 0.13 |
SPAG17 |
sperm associated antigen 17 |
83428 |
0.1 |
| chr17_19769997_19771171 | 0.13 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
646 |
0.74 |
| chr22_46474882_46475506 | 0.13 |
FLJ27365 |
hsa-mir-4763 |
998 |
0.36 |
| chr3_49169788_49170138 | 0.12 |
LAMB2 |
laminin, beta 2 (laminin S) |
490 |
0.65 |
| chr9_78589244_78589395 | 0.12 |
ENSG00000238826 |
. |
61950 |
0.14 |
| chr6_133067698_133067849 | 0.12 |
RP1-55C23.7 |
|
6041 |
0.13 |
| chr22_22011512_22012681 | 0.12 |
KB-1440D3.13 |
|
4127 |
0.1 |
| chrX_39964334_39965624 | 0.12 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr9_140500205_140501463 | 0.11 |
ARRDC1 |
arrestin domain containing 1 |
679 |
0.56 |
| chr3_50630246_50630397 | 0.10 |
HEMK1 |
HemK methyltransferase family member 1 |
15766 |
0.13 |
| chr19_45995836_45996758 | 0.10 |
RTN2 |
reticulon 2 |
228 |
0.87 |
| chr8_114450160_114450568 | 0.10 |
CSMD3 |
CUB and Sushi multiple domains 3 |
1036 |
0.64 |
| chr3_197044241_197044392 | 0.09 |
DLG1 |
discs, large homolog 1 (Drosophila) |
18145 |
0.15 |
| chr7_116312458_116312799 | 0.09 |
MET |
met proto-oncogene |
169 |
0.97 |
| chr10_95136415_95136839 | 0.09 |
MYOF |
myoferlin |
105324 |
0.06 |
| chr15_91285368_91285519 | 0.09 |
ENSG00000200677 |
. |
13099 |
0.14 |
| chr15_51633704_51634417 | 0.09 |
GLDN |
gliomedin |
234 |
0.91 |
| chr15_91383749_91383900 | 0.09 |
CTD-3094K11.1 |
|
870 |
0.48 |
| chr11_74352472_74352623 | 0.09 |
POLD3 |
polymerase (DNA-directed), delta 3, accessory subunit |
2888 |
0.24 |
| chr20_49137876_49138072 | 0.09 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
11054 |
0.16 |
| chr1_46337821_46337972 | 0.09 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
41364 |
0.17 |
| chr2_12842465_12842741 | 0.09 |
TRIB2 |
tribbles pseudokinase 2 |
14412 |
0.26 |
| chr13_67597561_67597712 | 0.08 |
PCDH9-AS4 |
PCDH9 antisense RNA 4 |
32618 |
0.21 |
| chr7_44923937_44924872 | 0.08 |
RP4-673M15.1 |
|
148 |
0.83 |
| chr14_53418107_53419061 | 0.08 |
FERMT2 |
fermitin family member 2 |
569 |
0.83 |
| chr22_46067402_46068067 | 0.08 |
ATXN10 |
ataxin 10 |
55 |
0.98 |
| chr8_53372834_53372985 | 0.08 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
50544 |
0.17 |
| chr3_134231000_134231151 | 0.08 |
CEP63 |
centrosomal protein 63kDa |
19616 |
0.18 |
| chr7_128171824_128173030 | 0.08 |
RP11-274B21.1 |
|
1279 |
0.43 |
| chr7_152549258_152549409 | 0.08 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
92419 |
0.08 |
| chr13_51483605_51485004 | 0.08 |
RNASEH2B |
ribonuclease H2, subunit B |
412 |
0.48 |
| chr6_109288525_109288676 | 0.08 |
RP11-787I22.3 |
|
31085 |
0.15 |
| chr4_160127685_160127927 | 0.08 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
60227 |
0.11 |
| chr20_36536600_36536751 | 0.08 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
5131 |
0.26 |
| chr1_157964729_157965124 | 0.08 |
KIRREL |
kin of IRRE like (Drosophila) |
1491 |
0.45 |
| chrX_38079311_38080259 | 0.08 |
SRPX |
sushi-repeat containing protein, X-linked |
367 |
0.75 |
| chr19_11009923_11010074 | 0.08 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
20607 |
0.11 |
| chr5_81701799_81702115 | 0.08 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
100791 |
0.08 |
| chr10_14287036_14287187 | 0.07 |
FRMD4A |
FERM domain containing 4A |
84990 |
0.09 |
| chr9_33523659_33524999 | 0.07 |
ANKRD18B |
ankyrin repeat domain 18B |
63 |
0.97 |
| chr1_210465594_210466382 | 0.07 |
HHAT |
hedgehog acyltransferase |
35608 |
0.17 |
| chr3_12200321_12200529 | 0.07 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
426 |
0.9 |
| chr1_203254106_203254257 | 0.07 |
BTG2 |
BTG family, member 2 |
20483 |
0.15 |
| chr3_11035286_11035437 | 0.07 |
SLC6A1 |
solute carrier family 6 (neurotransmitter transporter), member 1 |
947 |
0.65 |
| chr11_111411049_111411648 | 0.07 |
LAYN |
layilin |
36 |
0.96 |
| chr20_8381155_8381306 | 0.07 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
151858 |
0.04 |
| chr6_76645073_76645368 | 0.07 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
454 |
0.81 |
| chr16_57722748_57722899 | 0.07 |
CCDC135 |
coiled-coil domain containing 135 |
5882 |
0.14 |
| chrX_35937822_35938310 | 0.07 |
CXorf22 |
chromosome X open reading frame 22 |
215 |
0.96 |
| chr2_102908127_102908384 | 0.07 |
IL1RL1 |
interleukin 1 receptor-like 1 |
19707 |
0.18 |
| chr6_140783973_140784124 | 0.07 |
ENSG00000221336 |
. |
197802 |
0.03 |
| chr8_97657180_97658239 | 0.07 |
CPQ |
carboxypeptidase Q |
106 |
0.98 |
| chr20_49177863_49178105 | 0.07 |
ENSG00000201501 |
. |
1562 |
0.28 |
| chr1_205648469_205649627 | 0.07 |
SLC45A3 |
solute carrier family 45, member 3 |
539 |
0.74 |
| chr1_234630809_234631231 | 0.07 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
16171 |
0.21 |
| chr16_73098585_73098971 | 0.07 |
ZFHX3 |
zinc finger homeobox 3 |
5181 |
0.26 |
| chr1_144708589_144708740 | 0.07 |
RP11-640M9.2 |
|
93533 |
0.07 |
| chr14_63904706_63904968 | 0.06 |
ENSG00000264995 |
. |
24887 |
0.16 |
| chr12_114313370_114313521 | 0.06 |
RP11-780K2.1 |
|
56929 |
0.15 |
| chr22_46933754_46934840 | 0.06 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
1230 |
0.46 |
| chr11_131479044_131479195 | 0.06 |
NTM |
neurotrimin |
27214 |
0.2 |
| chr2_54350719_54350886 | 0.06 |
ACYP2 |
acylphosphatase 2, muscle type |
486 |
0.85 |
| chr10_3598463_3598925 | 0.06 |
RP11-184A2.3 |
|
194565 |
0.03 |
| chr12_52400775_52401965 | 0.06 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
646 |
0.62 |
| chr8_15646506_15646657 | 0.06 |
TUSC3 |
tumor suppressor candidate 3 |
165992 |
0.04 |
| chr2_201560727_201560878 | 0.06 |
AOX1 |
aldehyde oxidase 1 |
33172 |
0.15 |
| chr17_30438687_30438838 | 0.06 |
RP11-640N20.9 |
|
2382 |
0.21 |
| chr3_15469035_15469832 | 0.06 |
EAF1 |
ELL associated factor 1 |
328 |
0.51 |
| chr17_77019963_77021139 | 0.06 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
208 |
0.91 |
| chr7_45957495_45957807 | 0.06 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
626 |
0.73 |
| chr5_115298168_115298999 | 0.06 |
AQPEP |
Aminopeptidase Q |
392 |
0.86 |
| chr2_45179595_45180280 | 0.06 |
SIX3-AS1 |
SIX3 antisense RNA 1 |
10925 |
0.22 |
| chr10_73746466_73746617 | 0.06 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
22418 |
0.22 |
| chr6_5084934_5086227 | 0.06 |
PPP1R3G |
protein phosphatase 1, regulatory subunit 3G |
140 |
0.97 |
| chrX_65058405_65058556 | 0.06 |
RP6-159A1.3 |
|
161113 |
0.04 |
| chr8_77867156_77867509 | 0.06 |
ENSG00000266712 |
. |
11754 |
0.26 |
| chr2_228093116_228093390 | 0.06 |
AC097662.2 |
|
30661 |
0.17 |
| chr9_91653559_91653931 | 0.06 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
3307 |
0.28 |
| chr16_70470853_70471004 | 0.06 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
2063 |
0.21 |
| chr12_94242498_94242649 | 0.06 |
RP11-887P2.6 |
|
12630 |
0.2 |
| chr7_5732198_5732421 | 0.06 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
12217 |
0.2 |
| chr3_50328803_50329869 | 0.06 |
IFRD2 |
interferon-related developmental regulator 2 |
561 |
0.51 |
| chr2_173420797_173421943 | 0.06 |
AC093818.1 |
|
46 |
0.53 |
| chr4_187686525_187686676 | 0.06 |
FAT1 |
FAT atypical cadherin 1 |
38724 |
0.2 |
| chr17_1388273_1389008 | 0.06 |
MYO1C |
myosin IC |
21 |
0.97 |
| chr10_118896343_118896719 | 0.06 |
VAX1 |
ventral anterior homeobox 1 |
1036 |
0.51 |
| chr17_60885608_60885921 | 0.06 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
59 |
0.98 |
| chr2_26981186_26981361 | 0.06 |
SLC35F6 |
solute carrier family 35, member F6 |
5883 |
0.19 |
| chr3_129323468_129324020 | 0.06 |
PLXND1 |
plexin D1 |
1917 |
0.3 |
| chr3_134092531_134092774 | 0.06 |
AMOTL2 |
angiomotin like 2 |
13 |
0.98 |
| chr9_100697028_100697179 | 0.06 |
HEMGN |
hemogen |
10035 |
0.15 |
| chr10_93174739_93174920 | 0.05 |
HECTD2 |
HECT domain containing E3 ubiquitin protein ligase 2 |
4727 |
0.35 |
| chr3_64210682_64210890 | 0.05 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
345 |
0.9 |
| chr5_129353001_129353152 | 0.05 |
ENSG00000221562 |
. |
42492 |
0.18 |
| chrX_124337019_124337170 | 0.05 |
ENSG00000263886 |
. |
84531 |
0.11 |
| chr17_5412555_5412706 | 0.05 |
ENSG00000253071 |
. |
4960 |
0.15 |
| chr1_11072672_11073302 | 0.05 |
TARDBP |
TAR DNA binding protein |
246 |
0.86 |
| chrX_69674453_69675874 | 0.05 |
DLG3 |
discs, large homolog 3 (Drosophila) |
217 |
0.75 |
| chr15_99434112_99434263 | 0.05 |
IGF1R |
insulin-like growth factor 1 receptor |
617 |
0.73 |
| chr10_103734168_103734319 | 0.05 |
ENSG00000222430 |
. |
21782 |
0.16 |
| chr2_172266870_172267021 | 0.05 |
METTL8 |
methyltransferase like 8 |
23555 |
0.2 |
| chr21_36256728_36257172 | 0.05 |
RUNX1 |
runt-related transcription factor 1 |
2530 |
0.42 |
| chr10_15366222_15366373 | 0.05 |
FAM171A1 |
family with sequence similarity 171, member A1 |
46761 |
0.17 |
| chr11_121288254_121288502 | 0.05 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
34534 |
0.19 |
| chr3_157250943_157251353 | 0.05 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
247 |
0.94 |
| chr16_54970178_54971110 | 0.05 |
IRX5 |
iroquois homeobox 5 |
4660 |
0.36 |
| chr12_122667358_122668232 | 0.05 |
LRRC43 |
leucine rich repeat containing 43 |
128 |
0.95 |
| chr12_19018043_19018194 | 0.05 |
CAPZA3 |
capping protein (actin filament) muscle Z-line, alpha 3 |
127073 |
0.05 |
| chr16_68676411_68677035 | 0.05 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
2016 |
0.22 |
| chrX_14087477_14087628 | 0.05 |
ENSG00000264331 |
. |
6458 |
0.28 |
| chr3_50120540_50120691 | 0.05 |
RBM5 |
RNA binding motif protein 5 |
5726 |
0.16 |
| chr14_24836692_24836908 | 0.05 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
92 |
0.93 |
| chr14_101867418_101867569 | 0.05 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr5_40384613_40384764 | 0.05 |
ENSG00000265615 |
. |
64268 |
0.14 |
| chr13_99316705_99316856 | 0.05 |
ENSG00000243366 |
. |
7454 |
0.25 |
| chr1_159823893_159825316 | 0.05 |
C1orf204 |
chromosome 1 open reading frame 204 |
533 |
0.6 |
| chr22_26930460_26930611 | 0.05 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
6302 |
0.14 |
| chr16_48202466_48202617 | 0.05 |
RP11-3M1.1 |
|
1392 |
0.45 |
| chr1_92545604_92546569 | 0.05 |
BTBD8 |
BTB (POZ) domain containing 8 |
9 |
0.98 |
| chr7_16991756_16991907 | 0.05 |
AGR3 |
anterior gradient 3 |
70220 |
0.12 |
| chr17_60248884_60249035 | 0.05 |
ENSG00000207123 |
. |
49278 |
0.14 |
| chr2_121334219_121334381 | 0.05 |
ENSG00000201006 |
. |
74549 |
0.11 |
| chr11_7913917_7914068 | 0.05 |
ENSG00000252769 |
. |
3166 |
0.19 |
| chr14_70346080_70346697 | 0.05 |
SMOC1 |
SPARC related modular calcium binding 1 |
245 |
0.95 |
| chr15_67178585_67178736 | 0.05 |
SMAD6 |
SMAD family member 6 |
174625 |
0.03 |
| chr17_48278434_48278667 | 0.05 |
COL1A1 |
collagen, type I, alpha 1 |
443 |
0.73 |
| chr7_115117728_115117911 | 0.05 |
ENSG00000202377 |
. |
103677 |
0.08 |
| chr1_144983817_144983968 | 0.05 |
AL590452.1 |
Uncharacterized protein |
5417 |
0.16 |
| chr18_72778905_72779056 | 0.05 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
138488 |
0.05 |
| chr1_198859885_198860036 | 0.05 |
ENSG00000207759 |
. |
31678 |
0.2 |
| chr15_48937037_48938293 | 0.05 |
FBN1 |
fibrillin 1 |
253 |
0.95 |
| chr6_70576415_70577719 | 0.05 |
COL19A1 |
collagen, type XIX, alpha 1 |
604 |
0.84 |
| chr20_34043043_34043765 | 0.05 |
CEP250 |
centrosomal protein 250kDa |
141 |
0.87 |
| chr5_179495144_179495436 | 0.04 |
RNF130 |
ring finger protein 130 |
3431 |
0.26 |
| chr14_100204692_100204843 | 0.04 |
EML1 |
echinoderm microtubule associated protein like 1 |
681 |
0.73 |
| chr16_81534708_81535083 | 0.04 |
CMIP |
c-Maf inducing protein |
5941 |
0.3 |
| chr16_2077067_2078193 | 0.04 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
701 |
0.35 |
| chr1_206837249_206837437 | 0.04 |
ENSG00000252853 |
. |
17965 |
0.12 |
| chr19_1019836_1021109 | 0.04 |
TMEM259 |
transmembrane protein 259 |
373 |
0.62 |
| chr10_123871979_123872182 | 0.04 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
474 |
0.88 |
| chr10_131506190_131506341 | 0.04 |
RP11-109A6.2 |
|
6444 |
0.31 |
| chr1_161277026_161277213 | 0.04 |
MPZ |
myelin protein zero |
142 |
0.93 |
| chr22_33104594_33104745 | 0.04 |
ENSG00000251890 |
. |
73485 |
0.1 |
| chr10_104536560_104536809 | 0.04 |
WBP1L |
WW domain binding protein 1-like |
678 |
0.63 |
| chr7_66213885_66214036 | 0.04 |
RABGEF1 |
RAB guanine nucleotide exchange factor (GEF) 1 |
6916 |
0.16 |
| chr16_68440843_68441106 | 0.04 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
34813 |
0.11 |
| chr14_101292536_101293492 | 0.04 |
AL117190.2 |
|
2523 |
0.08 |
| chr6_16238952_16239854 | 0.04 |
GMPR |
guanosine monophosphate reductase |
592 |
0.74 |
| chr16_85558280_85558649 | 0.04 |
ENSG00000264203 |
. |
83366 |
0.08 |
| chr3_192568317_192568648 | 0.04 |
MB21D2 |
Mab-21 domain containing 2 |
67468 |
0.12 |
| chr14_60952017_60952553 | 0.04 |
C14orf39 |
chromosome 14 open reading frame 39 |
477 |
0.82 |
| chr4_145568031_145568374 | 0.04 |
HHIP |
hedgehog interacting protein |
878 |
0.49 |
| chr14_51835671_51835856 | 0.04 |
ENSG00000201820 |
. |
115022 |
0.05 |
| chr12_1628262_1628523 | 0.04 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
10665 |
0.24 |
| chr11_126286614_126287132 | 0.04 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
10196 |
0.19 |
| chr17_38336753_38336904 | 0.04 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
663 |
0.6 |
| chr10_15138786_15140121 | 0.04 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
34 |
0.62 |
| chr7_75830803_75830954 | 0.04 |
SRRM3 |
serine/arginine repetitive matrix 3 |
338 |
0.88 |
| chr22_31433016_31433229 | 0.04 |
ENSG00000240186 |
. |
22853 |
0.12 |
| chr12_31868549_31868829 | 0.04 |
AMN1 |
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
804 |
0.56 |
| chr8_146063147_146063298 | 0.04 |
ZNF7 |
zinc finger protein 7 |
8795 |
0.11 |
| chr19_13906290_13907522 | 0.04 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
632 |
0.39 |
| chr15_78799410_78800691 | 0.04 |
HYKK |
hydroxylysine kinase |
91 |
0.95 |
| chr7_121517277_121517428 | 0.04 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
3937 |
0.36 |
| chr5_35852042_35852193 | 0.04 |
IL7R |
interleukin 7 receptor |
680 |
0.72 |
| chr9_95788170_95788321 | 0.04 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
10903 |
0.19 |
| chr1_115753084_115753235 | 0.04 |
RP4-663N10.1 |
|
72496 |
0.11 |
| chr19_5036988_5037627 | 0.04 |
KDM4B |
lysine (K)-specific demethylase 4B |
20971 |
0.2 |
| chr7_134893359_134893623 | 0.04 |
WDR91 |
WD repeat domain 91 |
2160 |
0.24 |
| chr1_236958581_236959780 | 0.04 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
198 |
0.96 |
| chr2_9447040_9447191 | 0.04 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
100221 |
0.07 |
| chr6_49378033_49378231 | 0.04 |
MUT |
methylmalonyl CoA mutase |
52772 |
0.12 |
| chr21_44262526_44262677 | 0.04 |
WDR4 |
WD repeat domain 4 |
37040 |
0.15 |
| chr9_93881355_93881669 | 0.04 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
242665 |
0.02 |
| chr17_46177753_46179071 | 0.04 |
CBX1 |
chromobox homolog 1 |
148 |
0.91 |
| chr6_8433732_8434221 | 0.04 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
1740 |
0.54 |
| chr11_67807279_67808325 | 0.04 |
TCIRG1 |
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
1229 |
0.3 |
| chr3_128267814_128268156 | 0.04 |
C3orf27 |
chromosome 3 open reading frame 27 |
26944 |
0.16 |
| chr17_42071916_42072095 | 0.04 |
PYY |
peptide YY |
9832 |
0.1 |
| chr19_18716434_18716597 | 0.04 |
CRLF1 |
cytokine receptor-like factor 1 |
1145 |
0.26 |
| chr17_72738471_72738622 | 0.04 |
RAB37 |
RAB37, member RAS oncogene family |
5175 |
0.11 |
| chr9_38227227_38227501 | 0.04 |
ENSG00000238313 |
. |
36960 |
0.19 |
| chr19_4712831_4712982 | 0.04 |
DPP9 |
dipeptidyl-peptidase 9 |
1412 |
0.29 |
| chr19_8747658_8747869 | 0.04 |
ACTL9 |
actin-like 9 |
61409 |
0.09 |
| chr8_85095603_85095799 | 0.04 |
RALYL |
RALY RNA binding protein-like |
68 |
0.99 |
| chr4_99830188_99830339 | 0.04 |
RP11-571L19.7 |
|
19785 |
0.15 |
| chr14_101004037_101004188 | 0.04 |
BEGAIN |
brain-enriched guanylate kinase-associated |
10323 |
0.14 |
| chr13_32623224_32623468 | 0.04 |
FRY |
furry homolog (Drosophila) |
11655 |
0.23 |
| chr6_122931066_122932065 | 0.04 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
188 |
0.97 |
| chr5_159340244_159340395 | 0.04 |
ADRA1B |
adrenoceptor alpha 1B |
3471 |
0.33 |
| chr1_212810509_212810859 | 0.04 |
RP11-338C15.5 |
|
10571 |
0.14 |
| chr17_16190496_16190647 | 0.04 |
ENSG00000221355 |
. |
5243 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |