Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
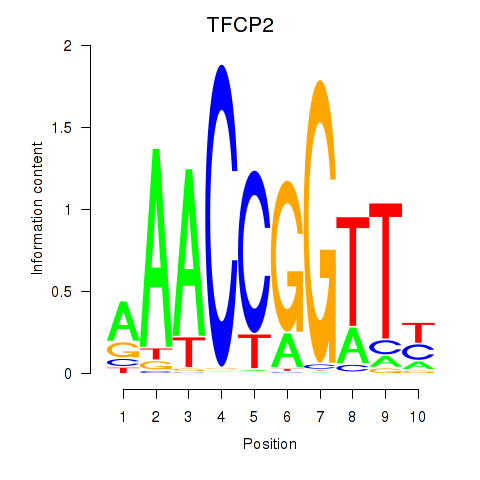
Results for TFCP2
Z-value: 1.30
Transcription factors associated with TFCP2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2
|
ENSG00000135457.5 | transcription factor CP2 |
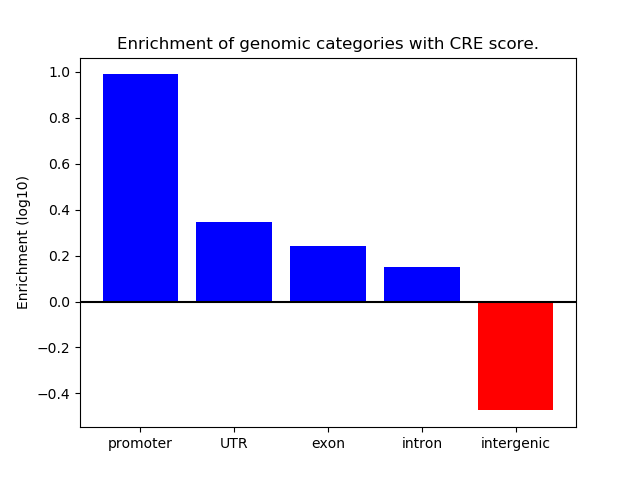
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_51565743_51565945 | TFCP2 | 732 | 0.557665 | 0.77 | 1.6e-02 | Click! |
| chr12_51567069_51567255 | TFCP2 | 236 | 0.890721 | -0.32 | 4.0e-01 | Click! |
| chr12_51566615_51566911 | TFCP2 | 68 | 0.958729 | -0.27 | 4.8e-01 | Click! |
| chr12_51563573_51563724 | TFCP2 | 2928 | 0.174409 | -0.22 | 5.7e-01 | Click! |
| chr12_51566002_51566548 | TFCP2 | 301 | 0.850201 | 0.21 | 5.8e-01 | Click! |
Activity of the TFCP2 motif across conditions
Conditions sorted by the z-value of the TFCP2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
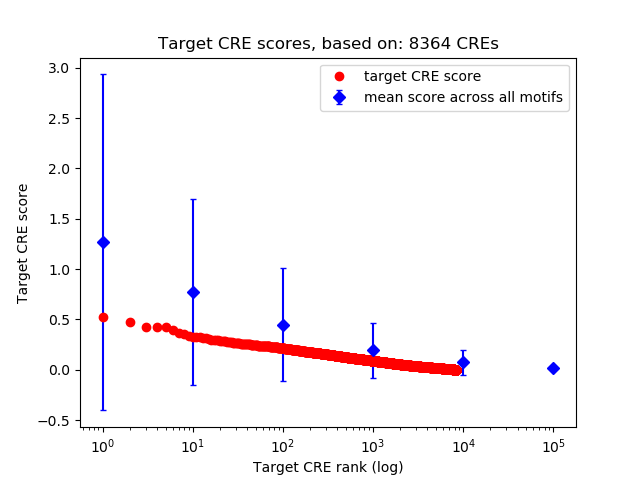
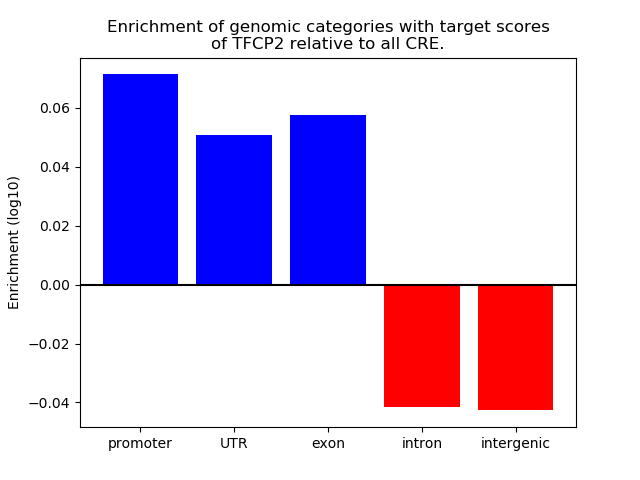
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_217929529_217929680 | 0.52 |
ENSG00000251849 |
. |
111617 |
0.06 |
| chr15_48936207_48936433 | 0.48 |
FBN1 |
fibrillin 1 |
1598 |
0.51 |
| chr3_106954545_106954751 | 0.43 |
ENSG00000220989 |
. |
129381 |
0.06 |
| chr4_16899159_16899373 | 0.43 |
LDB2 |
LIM domain binding 2 |
919 |
0.75 |
| chr22_40672488_40672756 | 0.42 |
TNRC6B |
trinucleotide repeat containing 6B |
11615 |
0.23 |
| chr18_56245541_56245920 | 0.39 |
RP11-126O1.2 |
|
21428 |
0.14 |
| chrX_34673275_34673426 | 0.36 |
TMEM47 |
transmembrane protein 47 |
2055 |
0.5 |
| chr4_55096991_55097208 | 0.36 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
610 |
0.82 |
| chr15_96870719_96871095 | 0.34 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
317 |
0.84 |
| chr7_15725092_15725754 | 0.33 |
MEOX2 |
mesenchyme homeobox 2 |
1014 |
0.63 |
| chr17_67137803_67138011 | 0.33 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
108 |
0.97 |
| chr7_19152487_19152712 | 0.33 |
AC003986.6 |
|
502 |
0.75 |
| chr9_18297085_18297314 | 0.32 |
ADAMTSL1 |
ADAMTS-like 1 |
176693 |
0.03 |
| chr1_26348142_26348482 | 0.32 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
41 |
0.95 |
| chr15_70994869_70995020 | 0.31 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
297 |
0.94 |
| chr3_52737763_52738047 | 0.30 |
GLT8D1 |
glycosyltransferase 8 domain containing 1 |
270 |
0.75 |
| chr11_119211431_119211655 | 0.30 |
C1QTNF5 |
C1q and tumor necrosis factor related protein 5 |
50 |
0.94 |
| chr14_55707492_55707643 | 0.30 |
FBXO34 |
F-box protein 34 |
30454 |
0.18 |
| chr5_158488269_158488591 | 0.30 |
EBF1 |
early B-cell factor 1 |
38271 |
0.17 |
| chr6_3389887_3390179 | 0.29 |
SLC22A23 |
solute carrier family 22, member 23 |
49619 |
0.15 |
| chr10_101767094_101767245 | 0.29 |
DNMBP |
dynamin binding protein |
2507 |
0.28 |
| chr1_114522141_114522332 | 0.29 |
OLFML3 |
olfactomedin-like 3 |
171 |
0.95 |
| chr5_129241672_129241829 | 0.28 |
CTC-575N7.1 |
|
140 |
0.95 |
| chr15_48936731_48936882 | 0.28 |
FBN1 |
fibrillin 1 |
1112 |
0.63 |
| chrX_34671991_34672311 | 0.28 |
TMEM47 |
transmembrane protein 47 |
3254 |
0.41 |
| chr8_13355358_13355509 | 0.28 |
DLC1 |
deleted in liver cancer 1 |
1317 |
0.52 |
| chr3_29321285_29321499 | 0.27 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
1081 |
0.59 |
| chr12_76422291_76422442 | 0.27 |
RP11-290L1.3 |
|
1908 |
0.31 |
| chr13_37492943_37493377 | 0.27 |
SMAD9 |
SMAD family member 9 |
1215 |
0.52 |
| chr19_55972195_55972559 | 0.27 |
ISOC2 |
isochorismatase domain containing 2 |
559 |
0.55 |
| chr17_15522812_15523031 | 0.27 |
CDRT1 |
CMT1A duplicated region transcript 1 |
95 |
0.95 |
| chr12_48395504_48395664 | 0.26 |
COL2A1 |
collagen, type II, alpha 1 |
2520 |
0.21 |
| chr2_102089965_102090218 | 0.26 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
646 |
0.8 |
| chr11_131780761_131781452 | 0.26 |
NTM |
neurotrimin |
209 |
0.95 |
| chr13_76054815_76055003 | 0.26 |
TBC1D4 |
TBC1 domain family, member 4 |
1341 |
0.51 |
| chr10_3666636_3666787 | 0.26 |
RP11-184A2.3 |
|
126548 |
0.06 |
| chr2_66667990_66668184 | 0.26 |
MEIS1 |
Meis homeobox 1 |
277 |
0.68 |
| chr16_11445129_11445310 | 0.26 |
RP11-485G7.6 |
|
2041 |
0.16 |
| chr2_216297601_216297993 | 0.26 |
FN1 |
fibronectin 1 |
2993 |
0.27 |
| chr11_118435857_118436109 | 0.25 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
378 |
0.76 |
| chr3_119180886_119181037 | 0.25 |
TMEM39A |
transmembrane protein 39A |
1502 |
0.36 |
| chr5_78760442_78760593 | 0.25 |
HOMER1 |
homer homolog 1 (Drosophila) |
48100 |
0.16 |
| chr7_46895478_46895629 | 0.25 |
AC011294.3 |
Uncharacterized protein |
158833 |
0.04 |
| chr14_61492437_61492630 | 0.25 |
SLC38A6 |
solute carrier family 38, member 6 |
17335 |
0.21 |
| chr12_119617823_119618008 | 0.25 |
HSPB8 |
heat shock 22kDa protein 8 |
520 |
0.73 |
| chr1_212734986_212735137 | 0.25 |
ATF3 |
activating transcription factor 3 |
3615 |
0.24 |
| chr12_50614114_50614771 | 0.25 |
RP3-405J10.4 |
|
1055 |
0.35 |
| chr15_60684258_60684446 | 0.25 |
ANXA2 |
annexin A2 |
856 |
0.71 |
| chr16_11085461_11085694 | 0.25 |
CLEC16A |
C-type lectin domain family 16, member A |
47139 |
0.1 |
| chr1_190447721_190448034 | 0.24 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
1118 |
0.69 |
| chr5_137390877_137391028 | 0.24 |
FAM13B |
family with sequence similarity 13, member B |
3302 |
0.16 |
| chr7_116035241_116035541 | 0.24 |
ENSG00000252672 |
. |
37877 |
0.14 |
| chr15_96883311_96883827 | 0.24 |
ENSG00000222651 |
. |
7079 |
0.16 |
| chr21_46629242_46629401 | 0.24 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
37764 |
0.14 |
| chr8_72460104_72460292 | 0.24 |
RP11-1102P16.1 |
Uncharacterized protein |
306 |
0.94 |
| chr5_14448576_14448727 | 0.24 |
TRIO |
trio Rho guanine nucleotide exchange factor |
39906 |
0.21 |
| chr13_36049575_36050388 | 0.24 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
851 |
0.43 |
| chr1_196622943_196623164 | 0.24 |
CFH |
complement factor H |
1867 |
0.41 |
| chr15_34786644_34787173 | 0.24 |
ENSG00000221065 |
. |
33664 |
0.14 |
| chr20_53092813_53093232 | 0.24 |
DOK5 |
docking protein 5 |
765 |
0.8 |
| chr6_85896028_85896266 | 0.24 |
NT5E |
5'-nucleotidase, ecto (CD73) |
263662 |
0.02 |
| chr19_11563038_11563189 | 0.24 |
ENSG00000238349 |
. |
13116 |
0.08 |
| chr14_55117285_55117487 | 0.24 |
SAMD4A |
sterile alpha motif domain containing 4A |
82749 |
0.09 |
| chr6_27516181_27516437 | 0.24 |
ENSG00000206671 |
. |
47882 |
0.13 |
| chr11_125929704_125929855 | 0.23 |
CDON |
cell adhesion associated, oncogene regulated |
2927 |
0.28 |
| chr5_79421582_79421788 | 0.23 |
CTC-458I2.2 |
|
2499 |
0.32 |
| chr1_164534283_164534434 | 0.23 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
2316 |
0.4 |
| chr17_32583150_32583439 | 0.23 |
AC005549.3 |
Uncharacterized protein |
988 |
0.32 |
| chr22_45898941_45899504 | 0.23 |
FBLN1 |
fibulin 1 |
296 |
0.92 |
| chr5_82770538_82771175 | 0.23 |
VCAN |
versican |
3112 |
0.37 |
| chr1_39733773_39734441 | 0.23 |
MACF1 |
microtubule-actin crosslinking factor 1 |
92 |
0.97 |
| chr12_72665965_72666270 | 0.23 |
ENSG00000236333 |
. |
189 |
0.69 |
| chr10_128994049_128994367 | 0.23 |
FAM196A |
family with sequence similarity 196, member A |
214 |
0.97 |
| chr18_56244732_56244930 | 0.23 |
RP11-126O1.2 |
|
20529 |
0.14 |
| chr12_105727681_105727958 | 0.23 |
C12orf75 |
chromosome 12 open reading frame 75 |
3173 |
0.24 |
| chr5_35129380_35129531 | 0.23 |
PRLR |
prolactin receptor |
991 |
0.64 |
| chr4_67431234_67431385 | 0.23 |
ENSG00000221563 |
. |
288767 |
0.01 |
| chr14_50089328_50089479 | 0.23 |
RP11-649E7.5 |
|
795 |
0.35 |
| chr18_55470424_55470575 | 0.23 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
172 |
0.96 |
| chr1_23066485_23066803 | 0.23 |
ENSG00000216157 |
. |
8760 |
0.17 |
| chr2_119532290_119532494 | 0.22 |
EN1 |
engrailed homeobox 1 |
72862 |
0.12 |
| chr4_26023554_26023967 | 0.22 |
SMIM20 |
small integral membrane protein 20 |
107829 |
0.07 |
| chr1_98510769_98511355 | 0.22 |
ENSG00000225206 |
. |
155 |
0.98 |
| chr7_128471351_128471889 | 0.22 |
FLNC |
filamin C, gamma |
1140 |
0.39 |
| chr1_240501387_240501653 | 0.22 |
ENSG00000252317 |
. |
3165 |
0.32 |
| chr4_19691164_19691362 | 0.22 |
ENSG00000251816 |
. |
507784 |
0.0 |
| chr12_110705323_110705474 | 0.22 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
13163 |
0.23 |
| chr11_133903795_133903999 | 0.22 |
JAM3 |
junctional adhesion molecule 3 |
34923 |
0.17 |
| chr10_95518290_95518441 | 0.22 |
LGI1 |
leucine-rich, glioma inactivated 1 |
687 |
0.76 |
| chr9_110811607_110811808 | 0.22 |
ENSG00000222459 |
. |
130448 |
0.05 |
| chr17_47210248_47210565 | 0.22 |
B4GALNT2 |
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
77 |
0.97 |
| chr12_40013401_40013886 | 0.22 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
90 |
0.98 |
| chr18_7460861_7461013 | 0.22 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
105843 |
0.07 |
| chr1_86045223_86045434 | 0.22 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
1116 |
0.4 |
| chr15_102156310_102156522 | 0.22 |
ENSG00000252614 |
. |
32099 |
0.15 |
| chrX_45626308_45626459 | 0.21 |
ENSG00000207725 |
. |
19853 |
0.22 |
| chr6_37663843_37664045 | 0.21 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
643 |
0.8 |
| chr12_125395024_125395175 | 0.21 |
UBC |
ubiquitin C |
3521 |
0.19 |
| chr1_87670295_87670446 | 0.21 |
ENSG00000221222 |
. |
53389 |
0.16 |
| chrX_64255707_64255858 | 0.21 |
ZC4H2 |
zinc finger, C4H2 domain containing |
1189 |
0.66 |
| chr10_62174775_62175056 | 0.21 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
25427 |
0.28 |
| chr1_215130950_215131319 | 0.21 |
KCNK2 |
potassium channel, subfamily K, member 2 |
48064 |
0.2 |
| chr3_105555914_105556177 | 0.21 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
31842 |
0.26 |
| chr10_92680601_92680752 | 0.21 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
357 |
0.84 |
| chr1_78474827_78475270 | 0.21 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
4510 |
0.17 |
| chr3_187870695_187870846 | 0.21 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
302 |
0.84 |
| chr2_75792595_75792746 | 0.21 |
EVA1A |
eva-1 homolog A (C. elegans) |
4178 |
0.2 |
| chr17_772969_773237 | 0.21 |
NXN |
nucleoredoxin |
5752 |
0.17 |
| chr1_117028958_117029109 | 0.21 |
ENSG00000200547 |
. |
33875 |
0.13 |
| chr16_86549558_86549709 | 0.21 |
FOXF1 |
forkhead box F1 |
5500 |
0.21 |
| chr14_77294740_77294891 | 0.21 |
C14orf166B |
chromosome 14 open reading frame 166B |
140 |
0.96 |
| chr12_78684646_78684797 | 0.21 |
NAV3 |
neuron navigator 3 |
91865 |
0.1 |
| chr10_31410381_31410532 | 0.21 |
ENSG00000263578 |
. |
55480 |
0.15 |
| chr14_42077252_42077403 | 0.21 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
291 |
0.93 |
| chr6_11230027_11230357 | 0.21 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
2699 |
0.33 |
| chr3_114378409_114378560 | 0.21 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
34692 |
0.22 |
| chr19_16493261_16493427 | 0.20 |
ENSG00000243745 |
. |
16426 |
0.13 |
| chr20_53239107_53239289 | 0.20 |
DOK5 |
docking protein 5 |
146941 |
0.05 |
| chr18_65662332_65662579 | 0.20 |
DSEL |
dermatan sulfate epimerase-like |
478238 |
0.01 |
| chr15_56206090_56206310 | 0.20 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
1652 |
0.45 |
| chr4_146052413_146052564 | 0.20 |
ENSG00000238713 |
. |
18313 |
0.17 |
| chrX_34673509_34673660 | 0.20 |
TMEM47 |
transmembrane protein 47 |
1821 |
0.53 |
| chr11_27603199_27603543 | 0.20 |
BDNF-AS |
BDNF antisense RNA |
58065 |
0.11 |
| chr6_151392043_151392194 | 0.20 |
RP1-292B18.3 |
|
14925 |
0.18 |
| chr19_30941733_30942003 | 0.20 |
ZNF536 |
zinc finger protein 536 |
78549 |
0.12 |
| chr4_177712658_177713139 | 0.20 |
VEGFC |
vascular endothelial growth factor C |
983 |
0.71 |
| chr6_35286420_35286751 | 0.20 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
9070 |
0.18 |
| chrX_51636554_51637686 | 0.20 |
MAGED1 |
melanoma antigen family D, 1 |
385 |
0.89 |
| chr4_41615256_41615763 | 0.20 |
LIMCH1 |
LIM and calponin homology domains 1 |
16 |
0.99 |
| chr5_92937754_92938000 | 0.20 |
ENSG00000237187 |
. |
16523 |
0.18 |
| chr4_39527980_39528193 | 0.20 |
UGDH |
UDP-glucose 6-dehydrogenase |
91 |
0.95 |
| chr11_76380846_76381073 | 0.20 |
LRRC32 |
leucine rich repeat containing 32 |
85 |
0.97 |
| chr1_60204181_60204332 | 0.20 |
ENSG00000266150 |
. |
5288 |
0.28 |
| chr19_36736141_36736420 | 0.20 |
ZNF565 |
zinc finger protein 565 |
879 |
0.5 |
| chr6_169652381_169652976 | 0.20 |
THBS2 |
thrombospondin 2 |
432 |
0.89 |
| chr15_37179413_37179956 | 0.20 |
ENSG00000212511 |
. |
34843 |
0.22 |
| chr9_102130860_102131451 | 0.20 |
ENSG00000222337 |
. |
84851 |
0.09 |
| chr8_129852489_129852640 | 0.20 |
ENSG00000221351 |
. |
20524 |
0.29 |
| chr9_119449393_119449768 | 0.20 |
TRIM32 |
tripartite motif containing 32 |
1 |
0.54 |
| chr8_96734481_96734632 | 0.20 |
ENSG00000223297 |
. |
43413 |
0.2 |
| chr17_57449049_57449286 | 0.19 |
ENSG00000263857 |
. |
5723 |
0.19 |
| chrX_135771582_135771733 | 0.19 |
CD40LG |
CD40 ligand |
41271 |
0.12 |
| chr9_1917275_1917426 | 0.19 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
97992 |
0.08 |
| chr9_82301184_82301335 | 0.19 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
18439 |
0.31 |
| chr5_33442964_33443296 | 0.19 |
TARS |
threonyl-tRNA synthetase |
1643 |
0.41 |
| chr22_31092194_31092345 | 0.19 |
OSBP2 |
oxysterol binding protein 2 |
1393 |
0.36 |
| chr10_63422698_63423173 | 0.19 |
C10orf107 |
chromosome 10 open reading frame 107 |
180 |
0.96 |
| chr7_107886376_107886580 | 0.19 |
NRCAM |
neuronal cell adhesion molecule |
2794 |
0.37 |
| chr7_18127374_18127525 | 0.19 |
HDAC9 |
histone deacetylase 9 |
877 |
0.66 |
| chr13_53173841_53174134 | 0.19 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
17618 |
0.22 |
| chr10_118547842_118547993 | 0.19 |
HSPA12A |
heat shock 70kDa protein 12A |
45832 |
0.13 |
| chr15_59622460_59622641 | 0.19 |
ENSG00000199512 |
. |
11066 |
0.14 |
| chr4_15471787_15471987 | 0.19 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
250 |
0.93 |
| chr10_375832_375983 | 0.19 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
441 |
0.85 |
| chr10_32153572_32153723 | 0.19 |
ARHGAP12 |
Rho GTPase activating protein 12 |
10539 |
0.28 |
| chr6_113994297_113994452 | 0.19 |
ENSG00000221559 |
. |
31661 |
0.2 |
| chr11_71750979_71751491 | 0.19 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
364 |
0.76 |
| chr13_36491458_36491923 | 0.19 |
DCLK1 |
doublecortin-like kinase 1 |
61911 |
0.15 |
| chr11_19092979_19093139 | 0.19 |
MRGPRX2 |
MAS-related GPR, member X2 |
10831 |
0.22 |
| chr17_48797815_48798020 | 0.19 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
909 |
0.47 |
| chr16_79605167_79605329 | 0.19 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
28551 |
0.25 |
| chr6_86159077_86159694 | 0.19 |
NT5E |
5'-nucleotidase, ecto (CD73) |
424 |
0.9 |
| chr9_179160_179373 | 0.19 |
CBWD1 |
COBW domain containing 1 |
194 |
0.94 |
| chrX_54947220_54948504 | 0.19 |
TRO |
trophinin |
41 |
0.97 |
| chr20_42087598_42088018 | 0.19 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
1240 |
0.39 |
| chr11_79148234_79148400 | 0.19 |
TENM4 |
teneurin transmembrane protein 4 |
3378 |
0.27 |
| chr10_4223311_4223462 | 0.19 |
ENSG00000207124 |
. |
333758 |
0.01 |
| chr1_117912075_117912374 | 0.18 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
2153 |
0.44 |
| chr4_108539974_108540125 | 0.18 |
PAPSS1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
101559 |
0.08 |
| chr19_14444112_14444508 | 0.18 |
CD97 |
CD97 molecule |
47658 |
0.1 |
| chr12_107352268_107352490 | 0.18 |
C12orf23 |
chromosome 12 open reading frame 23 |
1816 |
0.3 |
| chr2_44404368_44404519 | 0.18 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
8394 |
0.18 |
| chr15_49063488_49063639 | 0.18 |
RP11-485O10.2 |
|
11824 |
0.19 |
| chr2_150505079_150505230 | 0.18 |
ENSG00000207270 |
. |
37866 |
0.18 |
| chr8_54797720_54797871 | 0.18 |
RGS20 |
regulator of G-protein signaling 20 |
4341 |
0.21 |
| chr19_14605870_14606060 | 0.18 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
979 |
0.38 |
| chr6_22881696_22881893 | 0.18 |
ENSG00000207394 |
. |
243521 |
0.02 |
| chr2_42159815_42159972 | 0.18 |
C2orf91 |
chromosome 2 open reading frame 91 |
21050 |
0.21 |
| chr13_80707220_80707565 | 0.18 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
206402 |
0.03 |
| chr9_91093036_91093232 | 0.18 |
NXNL2 |
nucleoredoxin-like 2 |
56882 |
0.14 |
| chr20_3778661_3778819 | 0.18 |
CDC25B |
cell division cycle 25B |
1662 |
0.25 |
| chr10_33410829_33411028 | 0.18 |
ENSG00000263576 |
. |
23364 |
0.21 |
| chr7_43605854_43606005 | 0.18 |
ENSG00000252308 |
. |
12558 |
0.18 |
| chr3_161633215_161633436 | 0.18 |
OTOL1 |
otolin 1 |
418729 |
0.01 |
| chr3_139000860_139001011 | 0.18 |
MRPS22 |
mitochondrial ribosomal protein S22 |
61926 |
0.1 |
| chr6_90695271_90695484 | 0.18 |
ENSG00000222078 |
. |
15848 |
0.19 |
| chr6_85836289_85836440 | 0.18 |
NT5E |
5'-nucleotidase, ecto (CD73) |
323445 |
0.01 |
| chr6_99284839_99284990 | 0.18 |
POU3F2 |
POU class 3 homeobox 2 |
2334 |
0.45 |
| chr8_80525611_80525912 | 0.18 |
STMN2 |
stathmin-like 2 |
1791 |
0.51 |
| chr13_111862424_111862612 | 0.18 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
4882 |
0.27 |
| chr7_707964_708478 | 0.18 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
23210 |
0.18 |
| chr7_94030181_94030408 | 0.18 |
COL1A2 |
collagen, type I, alpha 2 |
6421 |
0.29 |
| chr22_40706773_40706982 | 0.18 |
ADSL |
adenylosuccinate lyase |
35630 |
0.14 |
| chr1_214620651_214620839 | 0.18 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
17401 |
0.26 |
| chr1_156677371_156677522 | 0.18 |
CRABP2 |
cellular retinoic acid binding protein 2 |
1838 |
0.18 |
| chr3_183966275_183966684 | 0.18 |
ALG3 |
ALG3, alpha-1,3- mannosyltransferase |
244 |
0.79 |
| chr2_192110869_192111020 | 0.18 |
MYO1B |
myosin IB |
65 |
0.98 |
| chr4_186064841_186065084 | 0.18 |
SLC25A4 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
567 |
0.76 |
| chr7_93925404_93925563 | 0.18 |
COL1A2 |
collagen, type I, alpha 2 |
98390 |
0.08 |
| chr1_33645945_33646389 | 0.18 |
TRIM62 |
tripartite motif containing 62 |
1100 |
0.55 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.6 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.1 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0060592 | mammary gland formation(GO:0060592) mammary placode formation(GO:0060596) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:2000053 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.6 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.3 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.0 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.3 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0014034 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.0 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0002248 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0032651 | regulation of interleukin-1 beta production(GO:0032651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |