Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
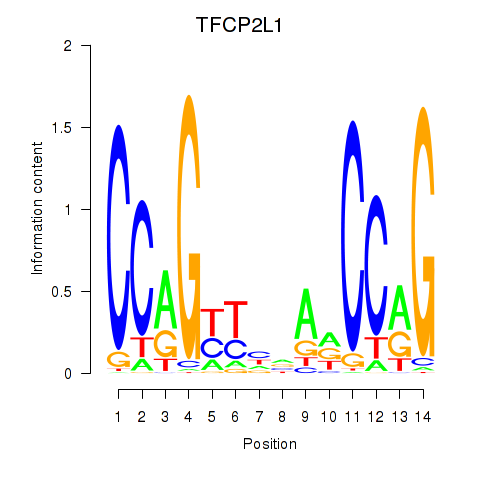
Results for TFCP2L1
Z-value: 0.61
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_122030038_122030189 | TFCP2L1 | 12670 | 0.291267 | 0.80 | 9.3e-03 | Click! |
| chr2_121969092_121969243 | TFCP2L1 | 73616 | 0.118802 | 0.78 | 1.3e-02 | Click! |
| chr2_121943565_121943716 | TFCP2L1 | 99143 | 0.078577 | 0.74 | 2.3e-02 | Click! |
| chr2_122040725_122040876 | TFCP2L1 | 1983 | 0.473434 | 0.68 | 4.3e-02 | Click! |
| chr2_122030438_122030589 | TFCP2L1 | 12270 | 0.292370 | 0.66 | 5.3e-02 | Click! |
Activity of the TFCP2L1 motif across conditions
Conditions sorted by the z-value of the TFCP2L1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
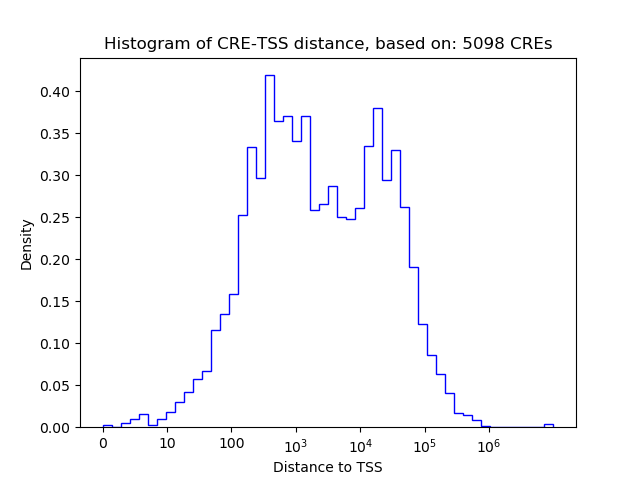
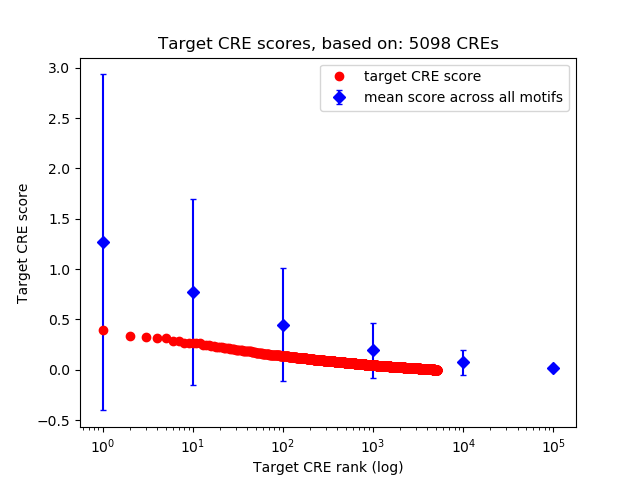
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_2143838_2144077 | 0.40 |
ENSG00000207805 |
. |
11482 |
0.11 |
| chr17_73718374_73718525 | 0.33 |
ITGB4 |
integrin, beta 4 |
872 |
0.43 |
| chr1_172360185_172360576 | 0.32 |
DNM3 |
dynamin 3 |
12222 |
0.16 |
| chr20_55204347_55205219 | 0.32 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr21_44753087_44753381 | 0.31 |
SIK1 |
salt-inducible kinase 1 |
93774 |
0.08 |
| chr2_227660537_227660688 | 0.29 |
IRS1 |
insulin receptor substrate 1 |
3863 |
0.25 |
| chr14_105332811_105333071 | 0.28 |
CEP170B |
centrosomal protein 170B |
1287 |
0.43 |
| chr11_131780111_131780399 | 0.27 |
NTM |
neurotrimin |
642 |
0.77 |
| chr12_54391177_54391923 | 0.26 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
981 |
0.22 |
| chr5_2752991_2753357 | 0.26 |
C5orf38 |
chromosome 5 open reading frame 38 |
795 |
0.6 |
| chr11_76495400_76495579 | 0.26 |
TSKU |
tsukushi, small leucine rich proteoglycan |
297 |
0.83 |
| chr16_765872_766413 | 0.26 |
METRN |
meteorin, glial cell differentiation regulator |
432 |
0.55 |
| chr21_46891372_46891858 | 0.25 |
COL18A1 |
collagen, type XVIII, alpha 1 |
16191 |
0.18 |
| chr1_110693132_110693795 | 0.25 |
SLC6A17 |
solute carrier family 6 (neutral amino acid transporter), member 17 |
355 |
0.81 |
| chr5_167646669_167646982 | 0.24 |
CTB-178M22.2 |
|
12537 |
0.22 |
| chr1_27189686_27189837 | 0.24 |
SFN |
stratifin |
128 |
0.93 |
| chr20_61467867_61468113 | 0.23 |
COL9A3 |
collagen, type IX, alpha 3 |
18827 |
0.11 |
| chr2_227661089_227661317 | 0.23 |
IRS1 |
insulin receptor substrate 1 |
3272 |
0.26 |
| chr12_6484544_6484695 | 0.23 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
24 |
0.57 |
| chr11_1896066_1896383 | 0.22 |
LSP1 |
lymphocyte-specific protein 1 |
1483 |
0.24 |
| chrX_48916544_48916695 | 0.22 |
CCDC120 |
coiled-coil domain containing 120 |
73 |
0.94 |
| chr8_42356526_42357223 | 0.21 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1887 |
0.34 |
| chr22_36461534_36461685 | 0.21 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
37136 |
0.19 |
| chr12_113859774_113860410 | 0.21 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr12_52540782_52540933 | 0.21 |
ENSG00000265804 |
. |
33148 |
0.09 |
| chr16_89630259_89630469 | 0.21 |
RPL13 |
ribosomal protein L13 |
1586 |
0.22 |
| chr17_75282403_75282641 | 0.21 |
SEPT9 |
septin 9 |
1451 |
0.48 |
| chr10_73758338_73758489 | 0.20 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
34290 |
0.18 |
| chr1_32045069_32045220 | 0.20 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
3005 |
0.19 |
| chr3_50193036_50193699 | 0.20 |
RP11-493K19.3 |
|
86 |
0.87 |
| chr11_1858026_1858177 | 0.20 |
SYT8 |
synaptotagmin VIII |
2067 |
0.16 |
| chr11_122049732_122050124 | 0.20 |
ENSG00000207994 |
. |
26912 |
0.16 |
| chr3_46428014_46428165 | 0.20 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
13928 |
0.14 |
| chr5_150968827_150968978 | 0.20 |
FAT2 |
FAT atypical cadherin 2 |
20397 |
0.17 |
| chr18_11689181_11689508 | 0.20 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
389 |
0.89 |
| chr9_132429402_132429765 | 0.19 |
PRRX2 |
paired related homeobox 2 |
1663 |
0.28 |
| chr5_135527631_135527866 | 0.19 |
TRPC7-AS1 |
TRPC7 antisense RNA 1 |
21988 |
0.18 |
| chr5_131593477_131594185 | 0.19 |
PDLIM4 |
PDZ and LIM domain 4 |
431 |
0.81 |
| chr20_33759810_33760395 | 0.19 |
PROCR |
protein C receptor, endothelial |
226 |
0.9 |
| chr6_16056248_16056399 | 0.19 |
MYLIP |
myosin regulatory light chain interacting protein |
73033 |
0.1 |
| chr18_77938309_77938600 | 0.19 |
AC139100.3 |
|
3109 |
0.25 |
| chr8_144896706_144897508 | 0.19 |
SCRIB |
scribbled planar cell polarity protein |
31 |
0.94 |
| chr17_878729_878960 | 0.18 |
NXN |
nucleoredoxin |
4166 |
0.16 |
| chr2_219858277_219858732 | 0.18 |
CRYBA2 |
crystallin, beta A2 |
361 |
0.74 |
| chr22_46980499_46980650 | 0.18 |
GRAMD4 |
GRAM domain containing 4 |
7548 |
0.22 |
| chr2_238804740_238804891 | 0.18 |
ENSG00000263723 |
. |
26268 |
0.17 |
| chr7_150814571_150814722 | 0.18 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
650 |
0.56 |
| chr13_114524710_114524921 | 0.18 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
6212 |
0.22 |
| chr4_187491209_187491867 | 0.17 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr16_75299159_75299651 | 0.17 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
292 |
0.87 |
| chr11_34642741_34642892 | 0.17 |
EHF |
ets homologous factor |
134 |
0.98 |
| chr10_64324269_64324457 | 0.17 |
ZNF365 |
zinc finger protein 365 |
44156 |
0.18 |
| chr12_116563914_116564400 | 0.17 |
ENSG00000207967 |
. |
22302 |
0.2 |
| chr14_105436556_105437076 | 0.16 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7878 |
0.16 |
| chr2_150186573_150186784 | 0.16 |
LYPD6 |
LY6/PLAUR domain containing 6 |
179 |
0.97 |
| chr2_86444196_86444456 | 0.16 |
MRPL35 |
mitochondrial ribosomal protein L35 |
17740 |
0.12 |
| chr22_37823258_37823437 | 0.16 |
RP1-63G5.5 |
|
133 |
0.53 |
| chr4_190731965_190732231 | 0.16 |
ENSG00000266024 |
. |
7725 |
0.28 |
| chr17_6459033_6459314 | 0.16 |
PITPNM3 |
PITPNM family member 3 |
607 |
0.7 |
| chr19_17953136_17953665 | 0.16 |
JAK3 |
Janus kinase 3 |
1856 |
0.22 |
| chr4_20256302_20256578 | 0.16 |
SLIT2 |
slit homolog 2 (Drosophila) |
103 |
0.98 |
| chr7_100876080_100876231 | 0.16 |
CLDN15 |
claudin 15 |
4886 |
0.11 |
| chrX_11682905_11683408 | 0.16 |
ARHGAP6 |
Rho GTPase activating protein 6 |
208 |
0.97 |
| chr15_37402857_37403086 | 0.16 |
MEIS2 |
Meis homeobox 2 |
9467 |
0.23 |
| chr5_131561351_131561783 | 0.16 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
1368 |
0.4 |
| chr5_61212914_61213065 | 0.16 |
RP11-2O17.2 |
|
218730 |
0.02 |
| chr6_13761461_13761614 | 0.15 |
MCUR1 |
mitochondrial calcium uniporter regulator 1 |
40040 |
0.15 |
| chr4_62067754_62067905 | 0.15 |
LPHN3 |
latrophilin 3 |
31 |
0.99 |
| chr2_225905702_225906810 | 0.15 |
DOCK10 |
dedicator of cytokinesis 10 |
903 |
0.7 |
| chr1_44030641_44030792 | 0.15 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
19969 |
0.18 |
| chr1_149908531_149908774 | 0.15 |
MTMR11 |
myotubularin related protein 11 |
139 |
0.92 |
| chr10_61469364_61469637 | 0.15 |
SLC16A9 |
solute carrier family 16, member 9 |
337 |
0.94 |
| chr14_55151103_55151261 | 0.15 |
SAMD4A |
sterile alpha motif domain containing 4A |
70369 |
0.12 |
| chr4_152462150_152462592 | 0.15 |
FAM160A1 |
family with sequence similarity 160, member A1 |
8207 |
0.21 |
| chr1_206685419_206685570 | 0.15 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
4615 |
0.16 |
| chr6_75911985_75912746 | 0.15 |
COL12A1 |
collagen, type XII, alpha 1 |
143 |
0.97 |
| chr9_116356768_116357019 | 0.15 |
RGS3 |
regulator of G-protein signaling 3 |
1127 |
0.54 |
| chr17_66596391_66596729 | 0.15 |
FAM20A |
family with sequence similarity 20, member A |
970 |
0.63 |
| chr2_69238294_69238445 | 0.15 |
ANTXR1 |
anthrax toxin receptor 1 |
1941 |
0.36 |
| chr8_145023500_145024036 | 0.15 |
PLEC |
plectin |
1276 |
0.29 |
| chr1_38100032_38100233 | 0.15 |
RSPO1 |
R-spondin 1 |
156 |
0.94 |
| chr1_3382753_3382904 | 0.15 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
259 |
0.93 |
| chr1_192776419_192777001 | 0.15 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
1461 |
0.52 |
| chr4_77561461_77561612 | 0.15 |
AC107072.2 |
|
2760 |
0.3 |
| chr11_7273101_7273359 | 0.14 |
SYT9 |
synaptotagmin IX |
49 |
0.98 |
| chr3_9745623_9746706 | 0.14 |
CPNE9 |
copine family member IX |
654 |
0.62 |
| chr5_98113748_98113899 | 0.14 |
RGMB |
repulsive guidance molecule family member b |
4484 |
0.25 |
| chr4_2066511_2066662 | 0.14 |
NAT8L |
N-acetyltransferase 8-like (GCN5-related, putative) |
5047 |
0.17 |
| chr1_220906357_220906642 | 0.14 |
MARC2 |
mitochondrial amidoxime reducing component 2 |
15112 |
0.19 |
| chr11_66104634_66104935 | 0.14 |
RIN1 |
Ras and Rab interactor 1 |
473 |
0.54 |
| chr19_12305803_12306072 | 0.14 |
ZNF136 |
zinc finger protein 136 |
32006 |
0.1 |
| chr7_127911991_127912224 | 0.14 |
ENSG00000221429 |
. |
17950 |
0.16 |
| chr3_117715779_117716482 | 0.14 |
LSAMP |
limbic system-associated membrane protein |
35 |
0.99 |
| chr2_241374262_241374834 | 0.14 |
GPC1 |
glypican 1 |
540 |
0.75 |
| chr8_94713195_94713677 | 0.14 |
FAM92A1 |
family with sequence similarity 92, member A1 |
326 |
0.7 |
| chr4_10097453_10097846 | 0.14 |
ENSG00000264931 |
. |
17333 |
0.14 |
| chr17_38334308_38334459 | 0.14 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
118 |
0.95 |
| chr9_35792725_35792965 | 0.14 |
NPR2 |
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
694 |
0.46 |
| chr17_74023240_74023391 | 0.14 |
EVPL |
envoplakin |
3 |
0.96 |
| chrX_118533808_118534027 | 0.14 |
SLC25A43 |
solute carrier family 25, member 43 |
574 |
0.64 |
| chr3_184258855_184259006 | 0.14 |
EIF2B5-AS1 |
EIF2B5 antisense RNA 1 |
15776 |
0.17 |
| chr17_55950685_55950914 | 0.14 |
CUEDC1 |
CUE domain containing 1 |
768 |
0.63 |
| chr3_126725554_126725705 | 0.14 |
PLXNA1 |
plexin A1 |
18192 |
0.27 |
| chr8_143869634_143869785 | 0.14 |
LY6D |
lymphocyte antigen 6 complex, locus D |
1701 |
0.2 |
| chr20_61002159_61002310 | 0.14 |
RBBP8NL |
RBBP8 N-terminal like |
355 |
0.83 |
| chr5_9544908_9545592 | 0.14 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
937 |
0.56 |
| chr6_3455960_3456202 | 0.14 |
SLC22A23 |
solute carrier family 22, member 23 |
196 |
0.97 |
| chr17_15686261_15686502 | 0.14 |
ENSG00000251829 |
. |
726 |
0.64 |
| chr10_127942305_127942518 | 0.14 |
ENSG00000222740 |
. |
108260 |
0.07 |
| chr2_28659437_28659588 | 0.13 |
PLB1 |
phospholipase B1 |
20500 |
0.17 |
| chr2_121943565_121943716 | 0.13 |
TFCP2L1 |
transcription factor CP2-like 1 |
99143 |
0.08 |
| chr9_85927301_85927452 | 0.13 |
FRMD3 |
FERM domain containing 3 |
495 |
0.81 |
| chr1_212415011_212415162 | 0.13 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
43793 |
0.13 |
| chr15_91500342_91500803 | 0.13 |
RCCD1 |
RCC1 domain containing 1 |
1546 |
0.18 |
| chr9_90812890_90813286 | 0.13 |
ENSG00000252299 |
. |
176096 |
0.03 |
| chr17_48133472_48134260 | 0.13 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
62 |
0.96 |
| chr17_5025740_5025978 | 0.13 |
ZNF232 |
zinc finger protein 232 |
538 |
0.63 |
| chr14_89506858_89507416 | 0.13 |
FOXN3 |
forkhead box N3 |
139951 |
0.05 |
| chr16_87241382_87241645 | 0.13 |
RP11-899L11.3 |
|
8008 |
0.23 |
| chr12_59309288_59309823 | 0.13 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1191 |
0.55 |
| chr20_30557048_30557199 | 0.13 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
1318 |
0.34 |
| chr17_37856067_37857084 | 0.13 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
242 |
0.86 |
| chr11_392408_392559 | 0.13 |
PKP3 |
plakophilin 3 |
131 |
0.92 |
| chr17_4902045_4902282 | 0.13 |
KIF1C |
kinesin family member 1C |
914 |
0.26 |
| chr9_98980169_98980538 | 0.13 |
ENSG00000265367 |
. |
17851 |
0.18 |
| chr9_130330127_130331202 | 0.13 |
FAM129B |
family with sequence similarity 129, member B |
703 |
0.67 |
| chr7_55120970_55121121 | 0.13 |
EGFR |
epidermal growth factor receptor |
34234 |
0.22 |
| chr2_175206758_175207367 | 0.13 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
4911 |
0.18 |
| chr17_11143475_11144119 | 0.13 |
SHISA6 |
shisa family member 6 |
783 |
0.71 |
| chr2_239776059_239776362 | 0.13 |
TWIST2 |
twist family bHLH transcription factor 2 |
19537 |
0.23 |
| chr2_153399155_153399498 | 0.13 |
FMNL2 |
formin-like 2 |
76766 |
0.12 |
| chr11_47561392_47561727 | 0.13 |
CELF1 |
CUGBP, Elav-like family member 1 |
13110 |
0.08 |
| chr18_59666418_59666569 | 0.13 |
RNF152 |
ring finger protein 152 |
105029 |
0.07 |
| chr2_151342010_151342200 | 0.12 |
RND3 |
Rho family GTPase 3 |
209 |
0.97 |
| chr7_101796468_101796619 | 0.12 |
ENSG00000252824 |
. |
40901 |
0.15 |
| chr11_19585849_19586032 | 0.12 |
ENSG00000265210 |
. |
10917 |
0.19 |
| chr2_219081816_219081967 | 0.12 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
27 |
0.96 |
| chr17_72426935_72427098 | 0.12 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
36 |
0.97 |
| chr21_37850808_37851450 | 0.12 |
CLDN14 |
claudin 14 |
1259 |
0.42 |
| chr10_73060971_73061122 | 0.12 |
SLC29A3 |
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
17969 |
0.18 |
| chr8_494094_494251 | 0.12 |
TDRP |
testis development related protein |
665 |
0.79 |
| chrX_56829451_56830094 | 0.12 |
ENSG00000204272 |
. |
74080 |
0.12 |
| chr6_35360609_35360760 | 0.12 |
PPARD |
peroxisome proliferator-activated receptor delta |
50282 |
0.11 |
| chr19_44284831_44285136 | 0.12 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
426 |
0.76 |
| chr10_60580536_60580825 | 0.12 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
27385 |
0.23 |
| chr13_102068554_102068730 | 0.12 |
NALCN |
sodium leak channel, non-selective |
64 |
0.98 |
| chr5_374526_374728 | 0.12 |
AHRR |
aryl-hydrocarbon receptor repressor |
30984 |
0.14 |
| chr1_110599711_110600002 | 0.12 |
RP4-773N10.4 |
|
1106 |
0.39 |
| chr10_128595169_128595405 | 0.12 |
DOCK1 |
dedicator of cytokinesis 1 |
1309 |
0.52 |
| chr1_210424867_210425148 | 0.12 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
17615 |
0.21 |
| chr15_74724718_74725429 | 0.12 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
928 |
0.5 |
| chr19_677691_677842 | 0.12 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
176 |
0.9 |
| chr1_152758757_152758908 | 0.12 |
LCE1E |
late cornified envelope 1E |
79 |
0.94 |
| chr10_3806458_3806609 | 0.12 |
RP11-184A2.2 |
|
3574 |
0.26 |
| chr1_159171046_159171197 | 0.12 |
CTA-134P22.2 |
|
1181 |
0.37 |
| chr12_41286154_41286409 | 0.12 |
CNTN1 |
contactin 1 |
15875 |
0.3 |
| chr1_156630677_156631393 | 0.12 |
RP11-284F21.7 |
|
181 |
0.89 |
| chr15_65714510_65715080 | 0.12 |
IGDCC4 |
immunoglobulin superfamily, DCC subclass, member 4 |
615 |
0.71 |
| chr11_63530349_63531105 | 0.12 |
ENSG00000264519 |
. |
34816 |
0.12 |
| chr6_15505544_15505788 | 0.12 |
DTNBP1 |
dystrobrevin binding protein 1 |
42927 |
0.19 |
| chr5_37839467_37839691 | 0.12 |
GDNF |
glial cell derived neurotrophic factor |
203 |
0.96 |
| chr3_62859805_62860096 | 0.12 |
CADPS |
Ca++-dependent secretion activator |
754 |
0.75 |
| chr15_75953728_75953957 | 0.12 |
SNX33 |
sorting nexin 33 |
11745 |
0.09 |
| chr17_79013898_79014087 | 0.12 |
BAIAP2 |
BAI1-associated protein 2 |
4189 |
0.15 |
| chr20_36846491_36846985 | 0.12 |
KIAA1755 |
KIAA1755 |
8883 |
0.19 |
| chr1_154786244_154786395 | 0.12 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
46000 |
0.11 |
| chr4_182470597_182470748 | 0.12 |
ENSG00000251742 |
. |
285589 |
0.01 |
| chr1_2141310_2141461 | 0.12 |
AL590822.1 |
Uncharacterized protein |
4235 |
0.14 |
| chr1_12675825_12676469 | 0.12 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1590 |
0.37 |
| chr3_49577030_49577181 | 0.12 |
BSN |
bassoon presynaptic cytomatrix protein |
14817 |
0.15 |
| chr4_55093927_55094291 | 0.11 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
1155 |
0.61 |
| chr2_236574591_236575244 | 0.11 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
3331 |
0.35 |
| chr14_75077914_75078284 | 0.11 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
676 |
0.71 |
| chr8_69242814_69243988 | 0.11 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr8_124550438_124550806 | 0.11 |
FBXO32 |
F-box protein 32 |
2824 |
0.28 |
| chr2_97636030_97636181 | 0.11 |
ENSG00000238760 |
. |
11016 |
0.14 |
| chr8_54794434_54794715 | 0.11 |
RGS20 |
regulator of G-protein signaling 20 |
1120 |
0.49 |
| chr15_75092311_75092506 | 0.11 |
CSK |
c-src tyrosine kinase |
2153 |
0.19 |
| chr20_60930372_60930523 | 0.11 |
RP11-157P1.5 |
|
2378 |
0.19 |
| chr20_35170676_35170845 | 0.11 |
MYL9 |
myosin, light chain 9, regulatory |
860 |
0.53 |
| chr9_71939475_71940274 | 0.11 |
FAM189A2 |
family with sequence similarity 189, member A2 |
386 |
0.91 |
| chr11_122071962_122072152 | 0.11 |
ENSG00000207994 |
. |
49041 |
0.12 |
| chr2_220139020_220139392 | 0.11 |
TUBA4A |
tubulin, alpha 4a |
3392 |
0.09 |
| chr13_111040556_111041009 | 0.11 |
ENSG00000238629 |
. |
25770 |
0.2 |
| chr7_145093241_145093392 | 0.11 |
ENSG00000200673 |
. |
546139 |
0.0 |
| chr6_143995099_143995343 | 0.11 |
PHACTR2 |
phosphatase and actin regulator 2 |
3881 |
0.32 |
| chr19_4374282_4374693 | 0.11 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5802 |
0.08 |
| chr1_16481335_16481929 | 0.11 |
RP11-276H7.2 |
|
74 |
0.93 |
| chr17_18281005_18281222 | 0.11 |
EVPLL |
envoplakin-like |
137 |
0.93 |
| chr3_40428700_40429091 | 0.11 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
159 |
0.96 |
| chr16_811279_811430 | 0.11 |
MSLN |
mesothelin |
256 |
0.77 |
| chr1_16508264_16508425 | 0.11 |
ARHGEF19-AS1 |
ARHGEF19 antisense RNA 1 |
16005 |
0.1 |
| chr22_43005089_43005553 | 0.11 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
5542 |
0.12 |
| chr19_20150080_20150295 | 0.11 |
ZNF682 |
zinc finger protein 682 |
65 |
0.97 |
| chrX_68063428_68063579 | 0.11 |
EFNB1 |
ephrin-B1 |
14663 |
0.3 |
| chr2_95742439_95743056 | 0.11 |
AC103563.9 |
|
23826 |
0.15 |
| chr1_98518942_98519575 | 0.11 |
ENSG00000225206 |
. |
7531 |
0.32 |
| chr11_70962306_70962531 | 0.11 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
1205 |
0.6 |
| chr20_48989140_48989291 | 0.11 |
ENSG00000244376 |
. |
56805 |
0.13 |
| chr20_45141972_45142630 | 0.11 |
ZNF334 |
zinc finger protein 334 |
103 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.3 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.2 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0048799 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0072179 | mesonephric tubule formation(GO:0072172) nephric duct formation(GO:0072179) |
| 0.0 | 0.0 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:0022011 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |