Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for TFEC_MITF_ARNTL_BHLHE41
Z-value: 0.59
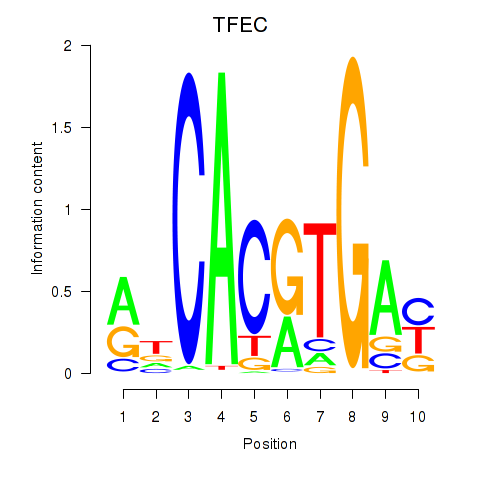
Transcription factors associated with TFEC_MITF_ARNTL_BHLHE41
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFEC
|
ENSG00000105967.11 | transcription factor EC |
|
MITF
|
ENSG00000187098.10 | melanocyte inducing transcription factor |
|
ARNTL
|
ENSG00000133794.13 | aryl hydrocarbon receptor nuclear translocator like |
|
BHLHE41
|
ENSG00000123095.5 | basic helix-loop-helix family member e41 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_13299425_13300004 | ARNTL | 282 | 0.944868 | 0.83 | 5.9e-03 | Click! |
| chr11_13326042_13326193 | ARNTL | 5416 | 0.284819 | 0.66 | 5.1e-02 | Click! |
| chr11_13298200_13298992 | ARNTL | 397 | 0.908274 | 0.64 | 6.4e-02 | Click! |
| chr11_13300075_13300637 | ARNTL | 924 | 0.699582 | 0.61 | 7.9e-02 | Click! |
| chr11_13238316_13238540 | ARNTL | 59771 | 0.146591 | 0.61 | 8.3e-02 | Click! |
| chr12_26278317_26278697 | BHLHE41 | 447 | 0.502305 | 0.45 | 2.3e-01 | Click! |
| chr12_26276607_26276969 | BHLHE41 | 1272 | 0.349442 | 0.32 | 4.1e-01 | Click! |
| chr12_26277242_26278053 | BHLHE41 | 413 | 0.758093 | 0.21 | 5.9e-01 | Click! |
| chr3_69887353_69887504 | MITF | 27935 | 0.220212 | 0.82 | 6.2e-03 | Click! |
| chr3_69814122_69814273 | MITF | 1235 | 0.533827 | -0.67 | 4.9e-02 | Click! |
| chr3_69811560_69811807 | MITF | 192 | 0.958954 | 0.53 | 1.4e-01 | Click! |
| chr3_69788021_69788388 | MITF | 382 | 0.904011 | 0.44 | 2.4e-01 | Click! |
| chr3_69698932_69699083 | MITF | 89579 | 0.089060 | -0.42 | 2.5e-01 | Click! |
| chr7_115668295_115668446 | TFEC | 2425 | 0.430817 | -0.62 | 7.3e-02 | Click! |
| chr7_115816578_115816729 | TFEC | 16703 | 0.213279 | 0.56 | 1.2e-01 | Click! |
| chr7_115806435_115806586 | TFEC | 6560 | 0.255445 | 0.46 | 2.1e-01 | Click! |
| chr7_115668575_115668726 | TFEC | 2145 | 0.458726 | -0.45 | 2.3e-01 | Click! |
| chr7_115665548_115665699 | TFEC | 5172 | 0.332938 | 0.44 | 2.4e-01 | Click! |
Activity of the TFEC_MITF_ARNTL_BHLHE41 motif across conditions
Conditions sorted by the z-value of the TFEC_MITF_ARNTL_BHLHE41 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
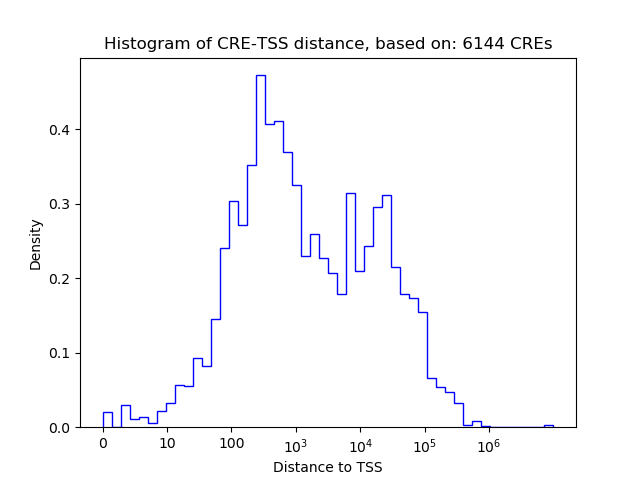
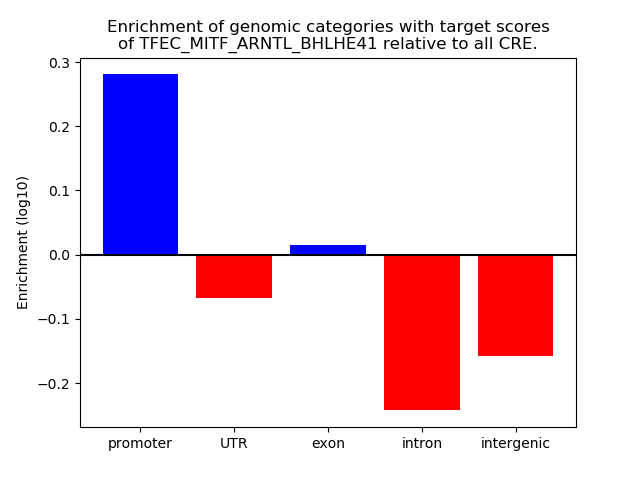
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_202896403_202896578 | 0.30 |
KLHL12 |
kelch-like family member 12 |
100 |
0.95 |
| chr19_4402510_4402661 | 0.29 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
74 |
0.93 |
| chr3_19988451_19988692 | 0.27 |
RAB5A |
RAB5A, member RAS oncogene family |
0 |
0.54 |
| chr1_154531335_154531531 | 0.25 |
UBE2Q1 |
ubiquitin-conjugating enzyme E2Q family member 1 |
324 |
0.81 |
| chr20_61315156_61315307 | 0.21 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
16066 |
0.11 |
| chr6_15897253_15897404 | 0.21 |
MYLIP |
myosin regulatory light chain interacting protein |
232028 |
0.02 |
| chr17_46646430_46646725 | 0.21 |
HOXB3 |
homeobox B3 |
4808 |
0.08 |
| chr19_10764777_10764928 | 0.20 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
85 |
0.95 |
| chr2_6868388_6868539 | 0.19 |
AC017076.5 |
|
105199 |
0.07 |
| chr2_202987834_202988283 | 0.19 |
AC079354.3 |
|
6277 |
0.17 |
| chr9_96793015_96793166 | 0.19 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
14 |
0.99 |
| chr1_27855501_27855753 | 0.18 |
RP1-159A19.4 |
|
3311 |
0.23 |
| chr17_39472322_39472473 | 0.18 |
KRTAP17-1 |
keratin associated protein 17-1 |
450 |
0.64 |
| chr10_135073626_135073923 | 0.18 |
MIR202HG |
MIR202 host gene (non-protein coding) |
12379 |
0.1 |
| chr11_63706516_63706697 | 0.17 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
114 |
0.94 |
| chr17_32906311_32906556 | 0.17 |
C17orf102 |
chromosome 17 open reading frame 102 |
45 |
0.97 |
| chr22_40406174_40406325 | 0.17 |
FAM83F |
family with sequence similarity 83, member F |
557 |
0.75 |
| chr13_49684124_49684410 | 0.17 |
FNDC3A |
fibronectin type III domain containing 3A |
182 |
0.96 |
| chr10_70480705_70480856 | 0.16 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
11 |
0.97 |
| chrX_102631036_102631748 | 0.16 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
57 |
0.97 |
| chr17_37271305_37271492 | 0.16 |
PLXDC1 |
plexin domain containing 1 |
7658 |
0.16 |
| chr9_126088906_126089175 | 0.15 |
CRB2 |
crumbs homolog 2 (Drosophila) |
29409 |
0.19 |
| chr2_126229593_126229744 | 0.15 |
ENSG00000201853 |
. |
343259 |
0.01 |
| chr7_644134_644285 | 0.15 |
AC147651.4 |
|
1387 |
0.35 |
| chr12_90103095_90103246 | 0.15 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
93 |
0.97 |
| chr8_28259515_28259714 | 0.15 |
ZNF395 |
zinc finger protein 395 |
604 |
0.66 |
| chr13_52377985_52378296 | 0.14 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
94 |
0.76 |
| chr15_63322236_63322431 | 0.14 |
TPM1 |
tropomyosin 1 (alpha) |
12498 |
0.18 |
| chr1_154193354_154193505 | 0.14 |
UBAP2L |
ubiquitin associated protein 2-like |
6 |
0.81 |
| chr14_62004309_62004586 | 0.14 |
RP11-47I22.1 |
|
7558 |
0.2 |
| chr3_184078420_184078571 | 0.14 |
CLCN2 |
chloride channel, voltage-sensitive 2 |
895 |
0.31 |
| chr7_150065813_150066064 | 0.14 |
REPIN1 |
replication initiator 1 |
22 |
0.52 |
| chr12_133262106_133262374 | 0.14 |
POLE |
polymerase (DNA directed), epsilon, catalytic subunit |
1674 |
0.22 |
| chr19_47200473_47200624 | 0.14 |
ENSG00000243560 |
. |
8223 |
0.09 |
| chr19_5681089_5681249 | 0.14 |
HSD11B1L |
hydroxysteroid (11-beta) dehydrogenase 1-like |
1 |
0.44 |
| chr9_98075387_98075786 | 0.13 |
FANCC |
Fanconi anemia, complementation group C |
3590 |
0.32 |
| chr12_85305306_85305778 | 0.13 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
261 |
0.96 |
| chr10_22609853_22610108 | 0.13 |
BMI1 |
BMI1 polycomb ring finger oncogene |
160 |
0.94 |
| chr17_79377761_79377965 | 0.13 |
ENSG00000266392 |
. |
3285 |
0.14 |
| chr10_104223217_104223368 | 0.13 |
TMEM180 |
transmembrane protein 180 |
2122 |
0.16 |
| chr5_139782206_139782432 | 0.13 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
813 |
0.3 |
| chr19_35939103_35939363 | 0.13 |
FFAR2 |
free fatty acid receptor 2 |
30 |
0.96 |
| chr10_3822916_3823697 | 0.13 |
KLF6 |
Kruppel-like factor 6 |
4079 |
0.25 |
| chr5_343263_343474 | 0.13 |
AHRR |
aryl-hydrocarbon receptor repressor |
275 |
0.91 |
| chr17_37910917_37911166 | 0.13 |
GRB7 |
growth factor receptor-bound protein 7 |
12536 |
0.11 |
| chr5_132759480_132759652 | 0.13 |
ENSG00000221287 |
. |
3832 |
0.29 |
| chr9_98278987_98279233 | 0.13 |
PTCH1 |
patched 1 |
137 |
0.95 |
| chr13_102106282_102106433 | 0.13 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
1339 |
0.56 |
| chr1_40283034_40283198 | 0.13 |
RP1-118J21.25 |
|
28468 |
0.11 |
| chr10_14050350_14050950 | 0.13 |
FRMD4A |
FERM domain containing 4A |
118 |
0.97 |
| chr19_10835608_10835759 | 0.13 |
DNM2 |
dynamin 2 |
6446 |
0.12 |
| chr3_158519414_158519896 | 0.12 |
MFSD1 |
major facilitator superfamily domain containing 1 |
1 |
0.54 |
| chr1_11866221_11866477 | 0.12 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
32 |
0.74 |
| chr4_148604275_148604426 | 0.12 |
PRMT10 |
protein arginine methyltransferase 10 (putative) |
926 |
0.6 |
| chr10_711598_711749 | 0.12 |
RP11-809C18.5 |
|
2175 |
0.26 |
| chr17_21156722_21156881 | 0.12 |
C17orf103 |
chromosome 17 open reading frame 103 |
79 |
0.97 |
| chr17_39261264_39261415 | 0.12 |
KRTAP4-9 |
keratin associated protein 4-9 |
245 |
0.77 |
| chr7_36010887_36011084 | 0.12 |
ENSG00000200446 |
. |
41111 |
0.17 |
| chr10_46090174_46090398 | 0.12 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
68 |
0.98 |
| chr17_66201588_66201894 | 0.12 |
AMZ2 |
archaelysin family metallopeptidase 2 |
41974 |
0.12 |
| chr1_56942848_56943117 | 0.12 |
ENSG00000223307 |
. |
99892 |
0.08 |
| chrX_72783084_72783245 | 0.12 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
120 |
0.98 |
| chr11_63933058_63933366 | 0.12 |
MACROD1 |
MACRO domain containing 1 |
321 |
0.79 |
| chr19_42757596_42757805 | 0.12 |
ERF |
Ets2 repressor factor |
351 |
0.71 |
| chr16_84401582_84401733 | 0.12 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
476 |
0.82 |
| chr16_29126642_29126901 | 0.12 |
CTB-134H23.3 |
|
8005 |
0.16 |
| chrX_100740137_100740443 | 0.12 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
2741 |
0.23 |
| chr15_31684340_31684641 | 0.12 |
KLF13 |
Kruppel-like factor 13 |
26133 |
0.26 |
| chr13_33000514_33000892 | 0.11 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
1448 |
0.44 |
| chr19_47523477_47523748 | 0.11 |
NPAS1 |
neuronal PAS domain protein 1 |
506 |
0.76 |
| chr6_13488651_13488802 | 0.11 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
832 |
0.5 |
| chr9_100684535_100684760 | 0.11 |
C9orf156 |
chromosome 9 open reading frame 156 |
122 |
0.95 |
| chr2_160372253_160373306 | 0.11 |
ENSG00000207117 |
. |
20078 |
0.23 |
| chr14_76446267_76446546 | 0.11 |
TGFB3 |
transforming growth factor, beta 3 |
930 |
0.62 |
| chr16_28306637_28306788 | 0.11 |
SBK1 |
SH3 domain binding kinase 1 |
2872 |
0.28 |
| chr14_72371815_72371966 | 0.11 |
RGS6 |
regulator of G-protein signaling 6 |
27266 |
0.24 |
| chr2_223916824_223917105 | 0.11 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
102 |
0.98 |
| chr11_66083004_66083200 | 0.11 |
CD248 |
CD248 molecule, endosialin |
1413 |
0.16 |
| chr17_21179296_21179691 | 0.11 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
8491 |
0.17 |
| chr4_2819558_2819838 | 0.11 |
SH3BP2 |
SH3-domain binding protein 2 |
214 |
0.94 |
| chr3_194992441_194992740 | 0.11 |
XXYLT1 |
xyloside xylosyltransferase 1 |
694 |
0.66 |
| chr1_224804905_224805156 | 0.11 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
1035 |
0.41 |
| chr2_234176635_234176786 | 0.11 |
ENSG00000252010 |
. |
7663 |
0.14 |
| chr16_23521764_23521915 | 0.11 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
24 |
0.97 |
| chr1_154908770_154909108 | 0.11 |
PMVK |
phosphomevalonate kinase |
528 |
0.6 |
| chr8_145114785_145114936 | 0.11 |
OPLAH |
5-oxoprolinase (ATP-hydrolysing) |
724 |
0.42 |
| chr1_42921657_42922194 | 0.11 |
ZMYND12 |
zinc finger, MYND-type containing 12 |
13 |
0.64 |
| chr2_46131469_46131620 | 0.11 |
PRKCE |
protein kinase C, epsilon |
96497 |
0.08 |
| chr15_34610151_34611295 | 0.11 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
257 |
0.87 |
| chr1_145611013_145611189 | 0.10 |
RNF115 |
ring finger protein 115 |
65 |
0.63 |
| chr7_34736_34914 | 0.10 |
FAM20C |
family with sequence similarity 20, member C |
158144 |
0.04 |
| chr9_117137299_117137450 | 0.10 |
AKNA |
AT-hook transcription factor |
1870 |
0.36 |
| chr3_126398343_126398521 | 0.10 |
TXNRD3 |
thioredoxin reductase 3 |
24434 |
0.17 |
| chr20_61916082_61916233 | 0.10 |
ENSG00000266104 |
. |
2003 |
0.18 |
| chr1_153508703_153508968 | 0.10 |
S100A6 |
S100 calcium binding protein A6 |
115 |
0.91 |
| chr1_182138463_182138614 | 0.10 |
ZNF648 |
zinc finger protein 648 |
107691 |
0.07 |
| chr6_43970266_43970484 | 0.10 |
RP5-1120P11.1 |
|
1011 |
0.46 |
| chr19_3367067_3367435 | 0.10 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
667 |
0.7 |
| chr11_27744105_27744362 | 0.10 |
BDNF |
brain-derived neurotrophic factor |
628 |
0.81 |
| chr14_20929873_20930024 | 0.10 |
TMEM55B |
transmembrane protein 55B |
177 |
0.87 |
| chr6_157469997_157470201 | 0.10 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
54 |
0.98 |
| chr1_6844402_6844649 | 0.10 |
RP11-312B8.1 |
|
378 |
0.72 |
| chr16_66914718_66914914 | 0.10 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
306 |
0.82 |
| chr15_72668375_72668880 | 0.10 |
HEXA-AS1 |
HEXA antisense RNA 1 |
173 |
0.51 |
| chr6_5026569_5027044 | 0.10 |
ENSG00000252419 |
. |
19529 |
0.19 |
| chr6_13030886_13031149 | 0.10 |
PHACTR1 |
phosphatase and actin regulator 1 |
22672 |
0.27 |
| chr22_33096549_33096700 | 0.10 |
ENSG00000251890 |
. |
65440 |
0.11 |
| chr17_58754842_58755035 | 0.10 |
BCAS3 |
breast carcinoma amplified sequence 3 |
124 |
0.96 |
| chr2_25149877_25150037 | 0.10 |
ADCY3 |
adenylate cyclase 3 |
7249 |
0.18 |
| chr13_80881448_80881599 | 0.10 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
32271 |
0.23 |
| chr18_74499301_74499473 | 0.10 |
ENSG00000252097 |
. |
13343 |
0.21 |
| chr9_132816560_132816877 | 0.10 |
GPR107 |
G protein-coupled receptor 107 |
733 |
0.67 |
| chr19_15236279_15236535 | 0.10 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
63 |
0.94 |
| chr6_7672816_7672976 | 0.10 |
BMP6 |
bone morphogenetic protein 6 |
54134 |
0.14 |
| chr15_96952055_96952206 | 0.10 |
AC087477.1 |
Uncharacterized protein |
47643 |
0.13 |
| chr2_54862437_54862588 | 0.10 |
AC093110.3 |
|
27417 |
0.18 |
| chr19_42900496_42900647 | 0.10 |
LIPE-AS1 |
LIPE antisense RNA 1 |
709 |
0.52 |
| chr6_42017736_42017887 | 0.10 |
CCND3 |
cyclin D3 |
284 |
0.63 |
| chr15_75248625_75249054 | 0.10 |
RPP25 |
ribonuclease P/MRP 25kDa subunit |
115 |
0.94 |
| chr16_73749020_73749171 | 0.10 |
C16orf47 |
chromosome 16 open reading frame 47 |
570749 |
0.0 |
| chr7_116593207_116593413 | 0.10 |
ST7 |
suppression of tumorigenicity 7 |
18 |
0.9 |
| chr22_38851841_38852002 | 0.10 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
716 |
0.57 |
| chr4_164471433_164471640 | 0.10 |
ENSG00000264535 |
. |
27092 |
0.19 |
| chr2_172963991_172964212 | 0.10 |
DLX2 |
distal-less homeobox 2 |
3527 |
0.26 |
| chr16_66914092_66914355 | 0.10 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
41 |
0.95 |
| chr1_7741272_7741423 | 0.10 |
CAMTA1 |
calmodulin binding transcription activator 1 |
55121 |
0.13 |
| chr1_8064348_8064644 | 0.09 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
11197 |
0.18 |
| chr17_47841796_47842381 | 0.09 |
FAM117A |
family with sequence similarity 117, member A |
559 |
0.72 |
| chr10_50969933_50970461 | 0.09 |
OGDHL |
oxoglutarate dehydrogenase-like |
172 |
0.97 |
| chr13_38230604_38230755 | 0.09 |
POSTN |
periostin, osteoblast specific factor |
57698 |
0.16 |
| chr17_68183563_68183714 | 0.09 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
17962 |
0.2 |
| chr15_44083848_44084237 | 0.09 |
SERF2 |
small EDRK-rich factor 2 |
2 |
0.94 |
| chr21_34401153_34401452 | 0.09 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
3059 |
0.26 |
| chr15_34788505_34788693 | 0.09 |
ENSG00000221065 |
. |
31973 |
0.15 |
| chr17_79455160_79455369 | 0.09 |
ENSG00000266077 |
. |
22871 |
0.08 |
| chr4_120222114_120222265 | 0.09 |
C4orf3 |
chromosome 4 open reading frame 3 |
113 |
0.97 |
| chr17_59018713_59018864 | 0.09 |
ENSG00000238799 |
. |
20466 |
0.18 |
| chr17_38708496_38708766 | 0.09 |
CCR7 |
chemokine (C-C motif) receptor 7 |
8634 |
0.17 |
| chr9_110495286_110496024 | 0.09 |
AL162389.1 |
Uncharacterized protein |
44764 |
0.15 |
| chr12_80083063_80083495 | 0.09 |
PAWR |
PRKC, apoptosis, WT1, regulator |
581 |
0.82 |
| chr9_131084429_131084680 | 0.09 |
TRUB2 |
TruB pseudouridine (psi) synthase family member 2 |
57 |
0.74 |
| chr2_119981485_119981702 | 0.09 |
STEAP3 |
STEAP family member 3, metalloreductase |
164 |
0.96 |
| chr12_123849561_123849862 | 0.09 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
321 |
0.85 |
| chr7_148637539_148638019 | 0.09 |
ENSG00000252310 |
. |
801 |
0.56 |
| chr11_33037172_33037593 | 0.09 |
DEPDC7 |
DEP domain containing 7 |
28 |
0.98 |
| chr14_104185073_104185224 | 0.09 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
3003 |
0.11 |
| chr3_97003394_97003545 | 0.09 |
EPHA6 |
EPH receptor A6 |
155005 |
0.04 |
| chr19_35615719_35615873 | 0.09 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
5529 |
0.1 |
| chr12_2185524_2185819 | 0.09 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
22942 |
0.16 |
| chr15_81316371_81316635 | 0.09 |
C15orf26 |
chromosome 15 open reading frame 26 |
17129 |
0.15 |
| chr12_52596811_52596962 | 0.09 |
KRT80 |
keratin 80 |
11102 |
0.12 |
| chr17_79031323_79031474 | 0.09 |
BAIAP2 |
BAI1-associated protein 2 |
17 |
0.97 |
| chr22_40742852_40743038 | 0.09 |
ADSL |
adenylosuccinate lyase |
409 |
0.84 |
| chr7_21985259_21985518 | 0.09 |
CDCA7L |
cell division cycle associated 7-like |
108 |
0.98 |
| chr19_6233769_6234001 | 0.09 |
CTC-503J8.4 |
|
25283 |
0.13 |
| chr11_280052_280224 | 0.09 |
NLRP6 |
NLR family, pyrin domain containing 6 |
1568 |
0.17 |
| chr10_29010987_29011704 | 0.09 |
ENSG00000201001 |
. |
38387 |
0.14 |
| chr13_30969133_30970071 | 0.09 |
HMGB1 |
high mobility group box 1 |
68782 |
0.12 |
| chr6_35744217_35744431 | 0.09 |
CLPSL2 |
colipase-like 2 |
68 |
0.96 |
| chr9_137437911_137438062 | 0.09 |
COL5A1 |
collagen, type V, alpha 1 |
95634 |
0.07 |
| chr12_66275134_66275285 | 0.08 |
RP11-366L20.2 |
Uncharacterized protein |
149 |
0.96 |
| chr9_102130860_102131451 | 0.08 |
ENSG00000222337 |
. |
84851 |
0.09 |
| chr4_164253766_164254020 | 0.08 |
NPY1R |
neuropeptide Y receptor Y1 |
121 |
0.97 |
| chr18_60384328_60384953 | 0.08 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
1957 |
0.36 |
| chr4_174451067_174451315 | 0.08 |
HAND2 |
heart and neural crest derivatives expressed 2 |
189 |
0.67 |
| chr17_62038790_62038941 | 0.08 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
11413 |
0.12 |
| chr11_106889276_106889822 | 0.08 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
299 |
0.95 |
| chr19_15218875_15219085 | 0.08 |
SYDE1 |
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
664 |
0.57 |
| chr7_100808865_100809016 | 0.08 |
VGF |
VGF nerve growth factor inducible |
66 |
0.94 |
| chr14_21852235_21852445 | 0.08 |
SUPT16H |
suppressor of Ty 16 homolog (S. cerevisiae) |
85 |
0.52 |
| chr19_36247080_36247289 | 0.08 |
HSPB6 |
heat shock protein, alpha-crystallin-related, B6 |
726 |
0.32 |
| chr17_65821514_65821703 | 0.08 |
BPTF |
bromodomain PHD finger transcription factor |
32 |
0.97 |
| chr9_33263612_33264186 | 0.08 |
BAG1 |
BCL2-associated athanogene |
658 |
0.51 |
| chr11_68607958_68608409 | 0.08 |
CPT1A |
carnitine palmitoyltransferase 1A (liver) |
1031 |
0.56 |
| chr19_890673_890824 | 0.08 |
MED16 |
mediator complex subunit 16 |
383 |
0.68 |
| chr6_133134294_133134903 | 0.08 |
RPS12 |
ribosomal protein S12 |
982 |
0.36 |
| chr15_96952221_96952372 | 0.08 |
AC087477.1 |
Uncharacterized protein |
47809 |
0.13 |
| chr20_23028362_23028696 | 0.08 |
THBD |
thrombomodulin |
1849 |
0.28 |
| chr7_2352113_2352318 | 0.08 |
SNX8 |
sorting nexin 8 |
1884 |
0.35 |
| chr20_48553017_48553241 | 0.08 |
RNF114 |
ring finger protein 114 |
181 |
0.92 |
| chr1_222885954_222886552 | 0.08 |
AIDA |
axin interactor, dorsalization associated |
273 |
0.56 |
| chr17_48450293_48450521 | 0.08 |
MRPL27 |
mitochondrial ribosomal protein L27 |
86 |
0.61 |
| chr8_143859070_143859321 | 0.08 |
LYNX1 |
Ly6/neurotoxin 1 |
31 |
0.95 |
| chr1_52831486_52831937 | 0.08 |
CC2D1B |
coiled-coil and C2 domain containing 1B |
140 |
0.95 |
| chr3_25825194_25825429 | 0.08 |
NGLY1 |
N-glycanase 1 |
269 |
0.8 |
| chr4_3529804_3529955 | 0.08 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
4407 |
0.19 |
| chr12_72148572_72148842 | 0.08 |
RAB21 |
RAB21, member RAS oncogene family |
53 |
0.98 |
| chr20_3661411_3661562 | 0.08 |
ADAM33 |
ADAM metallopeptidase domain 33 |
1264 |
0.38 |
| chr19_50379553_50379899 | 0.08 |
AKT1S1 |
AKT1 substrate 1 (proline-rich) |
220 |
0.74 |
| chr8_42434678_42434829 | 0.08 |
SMIM19 |
small integral membrane protein 19 |
33182 |
0.14 |
| chr12_69007685_69008546 | 0.08 |
RAP1B |
RAP1B, member of RAS oncogene family |
3310 |
0.27 |
| chr17_6915641_6915886 | 0.08 |
RNASEK |
ribonuclease, RNase K |
27 |
0.55 |
| chr17_80339190_80339423 | 0.08 |
UTS2R |
urotensin 2 receptor |
7153 |
0.1 |
| chr6_34499951_34500102 | 0.08 |
PACSIN1 |
protein kinase C and casein kinase substrate in neurons 1 |
17362 |
0.17 |
| chr3_148709208_148709359 | 0.08 |
GYG1 |
glycogenin 1 |
16 |
0.98 |
| chr16_71879481_71879729 | 0.08 |
ATXN1L |
ataxin 1-like |
289 |
0.47 |
| chr2_133427812_133428020 | 0.08 |
LYPD1 |
LY6/PLAUR domain containing 1 |
138 |
0.97 |
| chr20_13202499_13202944 | 0.08 |
ISM1 |
isthmin 1, angiogenesis inhibitor |
303 |
0.91 |
| chr16_54971124_54971492 | 0.08 |
IRX5 |
iroquois homeobox 5 |
5324 |
0.35 |
| chr1_1015177_1015464 | 0.08 |
RNF223 |
ring finger protein 223 |
5633 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |