Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
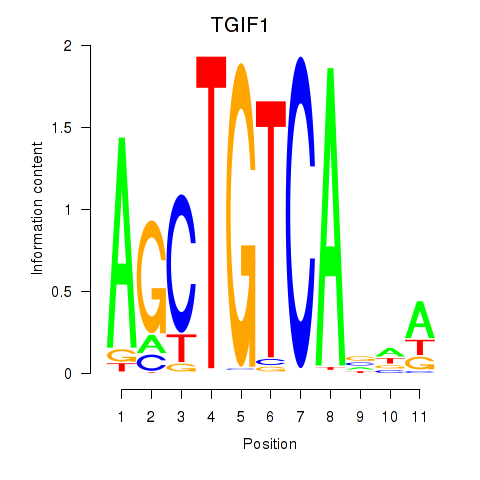
Results for TGIF1
Z-value: 0.52
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
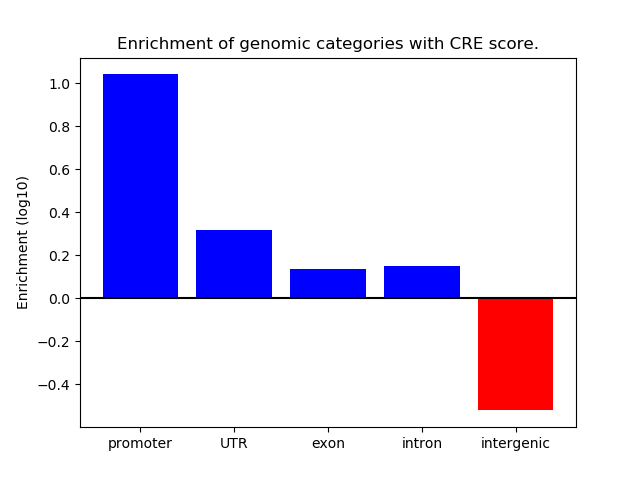
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_3462893_3463044 | TGIF1 | 7556 | 0.219731 | -0.62 | 7.5e-02 | Click! |
| chr18_3449680_3449975 | TGIF1 | 53 | 0.978836 | 0.58 | 1.0e-01 | Click! |
| chr18_3466935_3467086 | TGIF1 | 11598 | 0.208355 | -0.56 | 1.2e-01 | Click! |
| chr18_3451670_3451935 | TGIF1 | 80 | 0.976029 | 0.53 | 1.4e-01 | Click! |
| chr18_3448296_3449430 | TGIF1 | 366 | 0.884820 | 0.53 | 1.4e-01 | Click! |
Activity of the TGIF1 motif across conditions
Conditions sorted by the z-value of the TGIF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
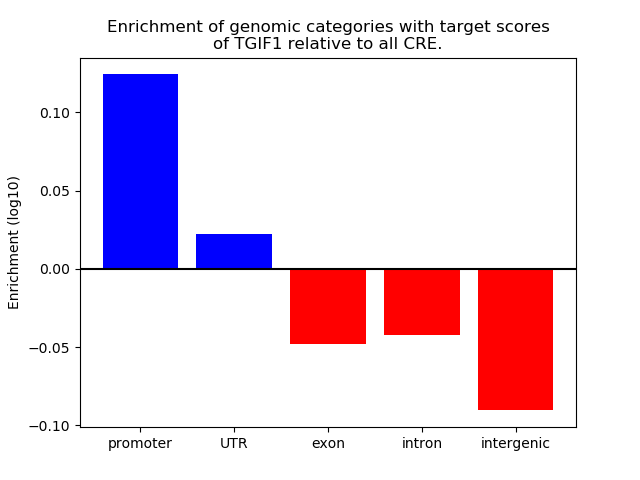
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_17587813_17587964 | 0.35 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
14255 |
0.14 |
| chr2_233925207_233925694 | 0.23 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
261 |
0.92 |
| chr2_143886988_143887259 | 0.22 |
ARHGAP15 |
Rho GTPase activating protein 15 |
240 |
0.95 |
| chr1_198608305_198608501 | 0.21 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
111 |
0.98 |
| chr15_93573821_93574103 | 0.21 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
21866 |
0.22 |
| chr11_63273553_63274069 | 0.20 |
LGALS12 |
lectin, galactoside-binding, soluble, 12 |
24 |
0.97 |
| chr1_160681036_160681588 | 0.20 |
CD48 |
CD48 molecule |
281 |
0.89 |
| chr17_4615541_4615774 | 0.19 |
ARRB2 |
arrestin, beta 2 |
1663 |
0.18 |
| chr1_47068584_47068735 | 0.19 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
1227 |
0.39 |
| chr8_1953776_1953973 | 0.17 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
31830 |
0.2 |
| chr10_73532392_73533248 | 0.17 |
C10orf54 |
chromosome 10 open reading frame 54 |
435 |
0.83 |
| chr17_56410837_56411117 | 0.16 |
MIR142 |
microRNA 142 |
1108 |
0.32 |
| chr7_157647247_157647398 | 0.16 |
AC011899.9 |
|
22 |
0.98 |
| chr6_133075391_133075542 | 0.16 |
RP1-55C23.7 |
|
1652 |
0.26 |
| chr15_58434934_58435085 | 0.16 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
3985 |
0.26 |
| chr12_25206187_25206540 | 0.16 |
LRMP |
lymphoid-restricted membrane protein |
689 |
0.71 |
| chr11_2322790_2323148 | 0.15 |
C11orf21 |
chromosome 11 open reading frame 21 |
174 |
0.62 |
| chr17_8316817_8316968 | 0.15 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
443 |
0.73 |
| chr15_94406450_94406737 | 0.15 |
ENSG00000222409 |
. |
33663 |
0.25 |
| chr17_62709857_62710178 | 0.15 |
RP13-104F24.1 |
|
44704 |
0.12 |
| chr12_94545481_94545632 | 0.14 |
PLXNC1 |
plexin C1 |
3057 |
0.29 |
| chrX_47488390_47488605 | 0.14 |
CFP |
complement factor properdin |
797 |
0.5 |
| chr1_169677205_169677356 | 0.14 |
SELL |
selectin L |
3559 |
0.24 |
| chr6_11382170_11382539 | 0.14 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
178 |
0.97 |
| chr3_14274334_14274485 | 0.14 |
XPC |
xeroderma pigmentosum, complementation group C |
54126 |
0.12 |
| chr11_128589269_128589524 | 0.14 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr12_93988367_93988869 | 0.14 |
SOCS2 |
suppressor of cytokine signaling 2 |
19784 |
0.17 |
| chr22_50523972_50524150 | 0.14 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
207 |
0.91 |
| chr7_37714542_37714738 | 0.14 |
GPR141 |
G protein-coupled receptor 141 |
8760 |
0.24 |
| chr16_57926919_57927200 | 0.13 |
CNGB1 |
cyclic nucleotide gated channel beta 1 |
8218 |
0.15 |
| chr6_35568962_35570321 | 0.13 |
ENSG00000212579 |
. |
49954 |
0.1 |
| chr10_94605706_94605908 | 0.13 |
EXOC6 |
exocyst complex component 6 |
2435 |
0.4 |
| chrX_71322682_71322935 | 0.13 |
RGAG4 |
retrotransposon gag domain containing 4 |
28870 |
0.14 |
| chr19_39826702_39826853 | 0.13 |
GMFG |
glia maturation factor, gamma |
48 |
0.94 |
| chr5_74964815_74965486 | 0.13 |
ENSG00000207333 |
. |
40274 |
0.14 |
| chr19_1068869_1069054 | 0.13 |
HMHA1 |
histocompatibility (minor) HA-1 |
1464 |
0.22 |
| chr12_25205029_25205380 | 0.13 |
LRMP |
lymphoid-restricted membrane protein |
1 |
0.98 |
| chr11_71854965_71855116 | 0.13 |
FOLR3 |
folate receptor 3 (gamma) |
8146 |
0.09 |
| chr7_130916444_130916595 | 0.13 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
53302 |
0.15 |
| chr2_170684384_170685373 | 0.12 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
860 |
0.53 |
| chr17_76259158_76259368 | 0.12 |
TMEM235 |
transmembrane protein 235 |
31141 |
0.11 |
| chr7_141646471_141646694 | 0.12 |
CLEC5A |
C-type lectin domain family 5, member A |
151 |
0.94 |
| chr9_75204897_75205048 | 0.12 |
TMC1 |
transmembrane channel-like 1 |
24644 |
0.24 |
| chr15_99499935_99500086 | 0.12 |
IGF1R |
insulin-like growth factor 1 receptor |
32181 |
0.16 |
| chr5_137449279_137449579 | 0.12 |
ENSG00000238605 |
. |
1564 |
0.23 |
| chr11_44588275_44588433 | 0.12 |
CD82 |
CD82 molecule |
1081 |
0.58 |
| chr2_158296405_158296556 | 0.12 |
CYTIP |
cytohesin 1 interacting protein |
554 |
0.73 |
| chr5_53886312_53886463 | 0.12 |
SNX18 |
sorting nexin 18 |
72794 |
0.12 |
| chr22_37639373_37639682 | 0.12 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
761 |
0.58 |
| chr1_236028992_236029750 | 0.12 |
LYST |
lysosomal trafficking regulator |
849 |
0.53 |
| chr19_13211050_13211262 | 0.12 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
2525 |
0.16 |
| chr3_172240144_172241280 | 0.12 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr8_134094409_134094685 | 0.12 |
SLA |
Src-like-adaptor |
20174 |
0.21 |
| chr4_37866428_37866579 | 0.12 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
26205 |
0.2 |
| chr6_140241181_140241350 | 0.12 |
ENSG00000252107 |
. |
238566 |
0.02 |
| chr1_40530106_40530257 | 0.12 |
PPT1 |
palmitoyl-protein thioesterase 1 |
17812 |
0.17 |
| chr13_108918883_108919034 | 0.12 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
3019 |
0.28 |
| chr3_16883275_16883542 | 0.11 |
PLCL2 |
phospholipase C-like 2 |
43044 |
0.19 |
| chr1_192128557_192128890 | 0.11 |
RGS18 |
regulator of G-protein signaling 18 |
1136 |
0.67 |
| chr17_43486558_43486834 | 0.11 |
ARHGAP27 |
Rho GTPase activating protein 27 |
1056 |
0.39 |
| chr6_6687519_6687926 | 0.11 |
LY86-AS1 |
LY86 antisense RNA 1 |
64718 |
0.14 |
| chr2_113599211_113599362 | 0.11 |
ENSG00000221541 |
. |
2602 |
0.24 |
| chr5_81074467_81075538 | 0.11 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr9_7653385_7653536 | 0.11 |
TMEM261 |
transmembrane protein 261 |
146607 |
0.05 |
| chr1_40804569_40804738 | 0.11 |
SMAP2 |
small ArfGAP2 |
5869 |
0.18 |
| chr15_58432158_58432309 | 0.11 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
1209 |
0.44 |
| chr6_160374740_160374891 | 0.11 |
IGF2R |
insulin-like growth factor 2 receptor |
15316 |
0.2 |
| chr21_36398861_36399526 | 0.11 |
RUNX1 |
runt-related transcription factor 1 |
22269 |
0.28 |
| chr3_67452818_67452969 | 0.11 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
125753 |
0.06 |
| chr12_105064873_105065024 | 0.11 |
ENSG00000264295 |
. |
79537 |
0.1 |
| chr9_134131290_134131650 | 0.11 |
FAM78A |
family with sequence similarity 78, member A |
14410 |
0.16 |
| chr19_2083107_2083981 | 0.11 |
MOB3A |
MOB kinase activator 3A |
1847 |
0.21 |
| chr15_93383088_93383239 | 0.11 |
ENSG00000264123 |
. |
16570 |
0.15 |
| chr1_207670300_207670451 | 0.11 |
CR1 |
complement component (3b/4b) receptor 1 (Knops blood group) |
689 |
0.7 |
| chr15_31558250_31558401 | 0.11 |
KLF13 |
Kruppel-like factor 13 |
60733 |
0.14 |
| chr13_108921982_108923111 | 0.11 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
302 |
0.91 |
| chr10_129846734_129847253 | 0.11 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1159 |
0.62 |
| chr1_192485875_192486153 | 0.11 |
ENSG00000221145 |
. |
25664 |
0.2 |
| chr5_139594597_139594748 | 0.11 |
CTB-131B5.2 |
|
14924 |
0.14 |
| chr3_183272411_183272703 | 0.11 |
KLHL6 |
kelch-like family member 6 |
920 |
0.55 |
| chr14_65511670_65511821 | 0.11 |
ENSG00000266531 |
. |
339 |
0.84 |
| chrX_9309324_9309596 | 0.11 |
TBL1X |
transducin (beta)-like 1X-linked |
121875 |
0.06 |
| chr2_68917176_68917327 | 0.10 |
ARHGAP25 |
Rho GTPase activating protein 25 |
20383 |
0.22 |
| chr10_106079197_106079348 | 0.10 |
RP11-127L20.5 |
|
7373 |
0.13 |
| chr1_149754304_149754780 | 0.10 |
FCGR1A |
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
296 |
0.78 |
| chr14_51394671_51394858 | 0.10 |
RP11-218E20.5 |
|
15820 |
0.16 |
| chr7_134836109_134836260 | 0.10 |
AC083862.1 |
Uncharacterized protein |
3268 |
0.17 |
| chr13_105792322_105792542 | 0.10 |
DAOA |
D-amino acid oxidase activator |
326160 |
0.01 |
| chr1_9690042_9690512 | 0.10 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
21513 |
0.15 |
| chr4_84406493_84406893 | 0.10 |
FAM175A |
family with sequence similarity 175, member A |
15 |
0.98 |
| chr2_60561577_60561728 | 0.10 |
ENSG00000200807 |
. |
50088 |
0.15 |
| chr9_92111480_92111784 | 0.10 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
715 |
0.59 |
| chr2_87834277_87834469 | 0.10 |
RP11-1399P15.1 |
|
56820 |
0.15 |
| chr16_2390604_2390763 | 0.10 |
ABCA3 |
ATP-binding cassette, sub-family A (ABC1), member 3 |
53 |
0.95 |
| chr1_12235343_12235565 | 0.10 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
8394 |
0.16 |
| chr1_150976139_150976311 | 0.10 |
FAM63A |
family with sequence similarity 63, member A |
2736 |
0.12 |
| chr6_133068019_133068170 | 0.10 |
RP1-55C23.7 |
|
5720 |
0.13 |
| chr6_7255590_7255764 | 0.10 |
RP11-69L16.4 |
|
20587 |
0.18 |
| chr6_2222017_2222664 | 0.10 |
GMDS |
GDP-mannose 4,6-dehydratase |
23586 |
0.28 |
| chr4_15255941_15256092 | 0.10 |
RP11-665G4.1 |
|
26432 |
0.23 |
| chrX_128914480_128914899 | 0.10 |
SASH3 |
SAM and SH3 domain containing 3 |
729 |
0.68 |
| chr20_52239578_52240391 | 0.10 |
ZNF217 |
zinc finger protein 217 |
14553 |
0.2 |
| chr8_126517925_126518109 | 0.10 |
ENSG00000266452 |
. |
61210 |
0.13 |
| chr10_35776313_35776464 | 0.10 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
117950 |
0.05 |
| chr9_126959092_126959243 | 0.10 |
NEK6 |
NIMA-related kinase 6 |
60718 |
0.11 |
| chr17_4617667_4617818 | 0.10 |
ARRB2 |
arrestin, beta 2 |
1142 |
0.27 |
| chr9_101899616_101899767 | 0.10 |
ENSG00000252942 |
. |
8557 |
0.19 |
| chr4_7061545_7061696 | 0.10 |
TADA2B |
transcriptional adaptor 2B |
6540 |
0.13 |
| chr17_4618141_4618477 | 0.10 |
ARRB2 |
arrestin, beta 2 |
575 |
0.53 |
| chr11_10622266_10622417 | 0.10 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
7597 |
0.18 |
| chr6_167142579_167142730 | 0.10 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
28745 |
0.23 |
| chr1_36912199_36912350 | 0.10 |
OSCP1 |
organic solute carrier partner 1 |
3606 |
0.17 |
| chr7_36231064_36231268 | 0.10 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
38289 |
0.15 |
| chr1_54871768_54871919 | 0.10 |
SSBP3 |
single stranded DNA binding protein 3 |
53 |
0.98 |
| chr2_238889883_238890034 | 0.10 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
8221 |
0.21 |
| chr11_65407442_65407916 | 0.10 |
SIPA1 |
signal-induced proliferation-associated 1 |
87 |
0.92 |
| chr8_38663476_38663627 | 0.10 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
583 |
0.73 |
| chr19_1856469_1856846 | 0.10 |
CTB-31O20.8 |
|
3592 |
0.1 |
| chr10_28822829_28823308 | 0.10 |
WAC |
WW domain containing adaptor with coiled-coil |
16 |
0.97 |
| chr4_40199803_40200006 | 0.10 |
RHOH |
ras homolog family member H |
1377 |
0.47 |
| chr11_1909549_1909755 | 0.10 |
C11orf89 |
chromosome 11 open reading frame 89 |
2432 |
0.16 |
| chr10_30831582_30831733 | 0.10 |
ENSG00000239744 |
. |
13176 |
0.24 |
| chr17_38374834_38374985 | 0.10 |
WIPF2 |
WAS/WASL interacting protein family, member 2 |
647 |
0.63 |
| chr6_108145054_108145346 | 0.09 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
316 |
0.93 |
| chr2_121096009_121096160 | 0.09 |
INHBB |
inhibin, beta B |
7635 |
0.26 |
| chr1_26453661_26453812 | 0.09 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
15396 |
0.1 |
| chr14_68283462_68283835 | 0.09 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
342 |
0.86 |
| chr3_186631546_186631697 | 0.09 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
16653 |
0.16 |
| chr7_102072016_102072236 | 0.09 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
1427 |
0.22 |
| chr6_11519933_11520084 | 0.09 |
ENSG00000202313 |
. |
16104 |
0.22 |
| chr16_81037806_81038351 | 0.09 |
CENPN |
centromere protein N |
2025 |
0.2 |
| chr19_14896102_14896264 | 0.09 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
6830 |
0.15 |
| chr19_49139505_49139656 | 0.09 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
1040 |
0.28 |
| chr20_58630648_58631293 | 0.09 |
C20orf197 |
chromosome 20 open reading frame 197 |
10 |
0.98 |
| chr1_90278526_90278677 | 0.09 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
7972 |
0.21 |
| chr19_36398415_36398818 | 0.09 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
533 |
0.58 |
| chr1_151170363_151170514 | 0.09 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
13 |
0.95 |
| chr11_129697989_129698558 | 0.09 |
TMEM45B |
transmembrane protein 45B |
12559 |
0.26 |
| chr12_51722623_51722774 | 0.09 |
BIN2 |
bridging integrator 2 |
4246 |
0.18 |
| chr6_106233724_106233875 | 0.09 |
ENSG00000200198 |
. |
118449 |
0.06 |
| chr10_129333006_129333157 | 0.09 |
NPS |
neuropeptide S |
14532 |
0.3 |
| chr9_101899309_101899497 | 0.09 |
ENSG00000252942 |
. |
8845 |
0.19 |
| chr11_77762153_77762304 | 0.09 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
4991 |
0.13 |
| chr3_169672312_169672463 | 0.09 |
RP11-379K17.4 |
|
11515 |
0.14 |
| chr4_185729074_185729225 | 0.09 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
456 |
0.81 |
| chr1_120935748_120935992 | 0.09 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
24 |
0.97 |
| chr22_17082347_17082639 | 0.09 |
ENSG00000221084 |
. |
2799 |
0.23 |
| chr19_13960913_13961064 | 0.09 |
ENSG00000207980 |
. |
13515 |
0.07 |
| chr6_29933308_29933483 | 0.09 |
HLA-A |
major histocompatibility complex, class I, A |
23069 |
0.14 |
| chr8_21754477_21754628 | 0.09 |
DOK2 |
docking protein 2, 56kDa |
16622 |
0.18 |
| chr17_29646389_29646953 | 0.09 |
CTD-2370N5.3 |
|
822 |
0.44 |
| chr3_35722538_35722689 | 0.09 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
121 |
0.97 |
| chr6_166667247_166667511 | 0.09 |
ENSG00000201292 |
. |
44488 |
0.14 |
| chr3_101405604_101405866 | 0.09 |
RPL24 |
ribosomal protein L24 |
109 |
0.94 |
| chr14_95927410_95927561 | 0.09 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
4959 |
0.23 |
| chr1_89591826_89592005 | 0.09 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
118 |
0.97 |
| chr13_41884953_41885353 | 0.09 |
NAA16 |
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
188 |
0.95 |
| chrX_48793999_48794379 | 0.09 |
PIM2 |
pim-2 oncogene |
17888 |
0.08 |
| chr20_58676178_58676329 | 0.09 |
C20orf197 |
chromosome 20 open reading frame 197 |
45273 |
0.17 |
| chr16_23864221_23864482 | 0.09 |
PRKCB |
protein kinase C, beta |
15807 |
0.23 |
| chr5_142985423_142985638 | 0.09 |
CTB-57H20.1 |
|
337 |
0.92 |
| chr6_119634479_119634630 | 0.09 |
ENSG00000200732 |
. |
13892 |
0.25 |
| chr8_142219697_142219848 | 0.09 |
DENND3 |
DENN/MADD domain containing 3 |
17342 |
0.15 |
| chr2_68477764_68478263 | 0.09 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
267 |
0.87 |
| chr11_10626007_10626277 | 0.09 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
11398 |
0.17 |
| chr11_2324787_2325062 | 0.09 |
TSPAN32 |
tetraspanin 32 |
826 |
0.45 |
| chr12_108909076_108909295 | 0.09 |
FICD |
FIC domain containing |
92 |
0.96 |
| chr8_53851786_53851937 | 0.09 |
NPBWR1 |
neuropeptides B/W receptor 1 |
870 |
0.7 |
| chr19_54713150_54713301 | 0.09 |
RPS9 |
ribosomal protein S9 |
8197 |
0.07 |
| chr6_42013197_42013548 | 0.09 |
CCND3 |
cyclin D3 |
3052 |
0.21 |
| chr12_117073537_117073765 | 0.08 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
59995 |
0.13 |
| chr1_36621953_36622250 | 0.08 |
MAP7D1 |
MAP7 domain containing 1 |
298 |
0.87 |
| chr17_29640754_29641122 | 0.08 |
EVI2B |
ecotropic viral integration site 2B |
164 |
0.93 |
| chr5_10445338_10445751 | 0.08 |
ROPN1L |
rhophilin associated tail protein 1-like |
3556 |
0.18 |
| chr8_48515171_48515322 | 0.08 |
SPIDR |
scaffolding protein involved in DNA repair |
56980 |
0.12 |
| chr8_126446403_126446861 | 0.08 |
TRIB1 |
tribbles pseudokinase 1 |
844 |
0.67 |
| chr12_4312868_4313019 | 0.08 |
CCND2 |
cyclin D2 |
69995 |
0.1 |
| chr4_129732983_129734245 | 0.08 |
JADE1 |
jade family PHD finger 1 |
615 |
0.83 |
| chr5_55884070_55884221 | 0.08 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
17914 |
0.22 |
| chr10_134385473_134385624 | 0.08 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
33905 |
0.18 |
| chr7_37389906_37390077 | 0.08 |
ENSG00000222869 |
. |
756 |
0.67 |
| chr17_73682901_73683052 | 0.08 |
RP11-474I11.7 |
|
2097 |
0.17 |
| chr13_50202297_50202533 | 0.08 |
ARL11 |
ADP-ribosylation factor-like 11 |
20 |
0.98 |
| chr11_2468512_2468676 | 0.08 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
2373 |
0.23 |
| chr14_73932367_73932610 | 0.08 |
ENSG00000251393 |
. |
3359 |
0.19 |
| chr8_95956511_95956662 | 0.08 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
3385 |
0.17 |
| chr2_54657230_54657381 | 0.08 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
26117 |
0.21 |
| chr1_116293768_116293919 | 0.08 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
17492 |
0.24 |
| chr9_132830835_132830986 | 0.08 |
GPR107 |
G protein-coupled receptor 107 |
14925 |
0.18 |
| chr4_6918537_6918793 | 0.08 |
TBC1D14 |
TBC1 domain family, member 14 |
6690 |
0.19 |
| chr11_77167116_77167267 | 0.08 |
DKFZP434E1119 |
|
17225 |
0.19 |
| chr15_43531850_43532099 | 0.08 |
ENSG00000202211 |
. |
7904 |
0.13 |
| chr17_8076861_8077643 | 0.08 |
ENSG00000200463 |
. |
347 |
0.7 |
| chr20_48962765_48962916 | 0.08 |
ENSG00000244376 |
. |
83180 |
0.09 |
| chr12_54693479_54693635 | 0.08 |
NFE2 |
nuclear factor, erythroid 2 |
1242 |
0.21 |
| chr5_10635367_10635573 | 0.08 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
7133 |
0.25 |
| chr5_163147994_163148145 | 0.08 |
ENSG00000251998 |
. |
75102 |
0.12 |
| chr16_85793045_85793254 | 0.08 |
C16orf74 |
chromosome 16 open reading frame 74 |
8414 |
0.11 |
| chr12_15113710_15114247 | 0.08 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
222 |
0.92 |
| chr19_50269466_50269673 | 0.08 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
656 |
0.43 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.0 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0032747 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0032490 | detection of molecule of bacterial origin(GO:0032490) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |