Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
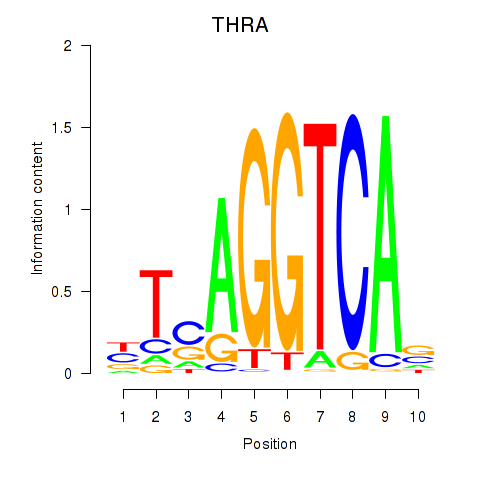
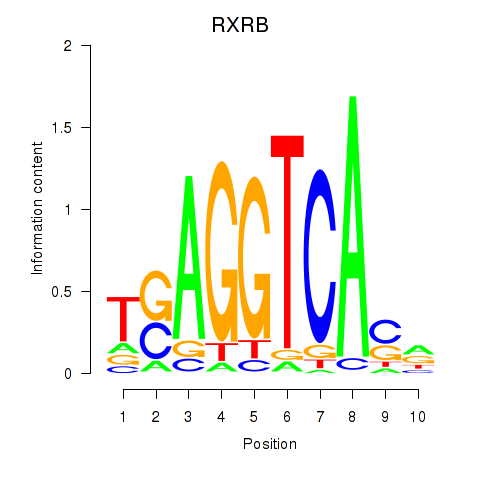
Results for THRA_RXRB
Z-value: 0.80
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
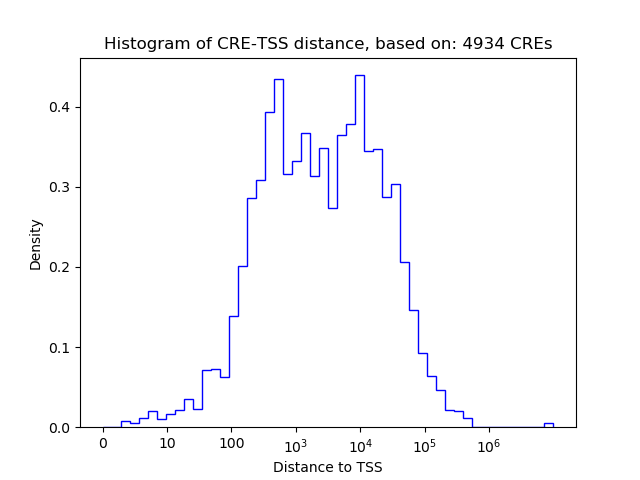
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_38214670_38214853 | THRA | 218 | 0.876638 | -0.77 | 1.5e-02 | Click! |
| chr17_38215195_38215346 | THRA | 727 | 0.511622 | -0.66 | 5.1e-02 | Click! |
| chr17_38224700_38225305 | THRA | 154 | 0.925872 | 0.66 | 5.4e-02 | Click! |
| chr17_38228617_38228812 | THRA | 370 | 0.788671 | -0.63 | 7.1e-02 | Click! |
| chr17_38221376_38221734 | THRA | 2403 | 0.180628 | -0.62 | 7.7e-02 | Click! |
Activity of the THRA_RXRB motif across conditions
Conditions sorted by the z-value of the THRA_RXRB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_134086172_134086360 | 0.61 |
SLA |
Src-like-adaptor |
13663 |
0.23 |
| chr13_114428625_114429103 | 0.61 |
TMEM255B |
transmembrane protein 255B |
33352 |
0.18 |
| chr16_28306637_28306788 | 0.61 |
SBK1 |
SH3 domain binding kinase 1 |
2872 |
0.28 |
| chr1_206732914_206733127 | 0.60 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2527 |
0.25 |
| chr6_154566868_154567140 | 0.54 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
986 |
0.71 |
| chr3_50646296_50646682 | 0.51 |
CISH |
cytokine inducible SH2-containing protein |
2714 |
0.2 |
| chr17_8867884_8868296 | 0.49 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
934 |
0.65 |
| chr3_183273082_183273407 | 0.49 |
KLHL6 |
kelch-like family member 6 |
233 |
0.92 |
| chr18_43267753_43268033 | 0.46 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
21786 |
0.17 |
| chr5_81074467_81075538 | 0.45 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr5_150593652_150593827 | 0.44 |
GM2A |
GM2 ganglioside activator |
2028 |
0.31 |
| chr6_128221812_128222507 | 0.44 |
THEMIS |
thymocyte selection associated |
56 |
0.99 |
| chr14_22964891_22965136 | 0.43 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
8842 |
0.1 |
| chr11_104904746_104905919 | 0.43 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
508 |
0.77 |
| chr2_68963054_68963510 | 0.40 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1268 |
0.56 |
| chr20_827167_827339 | 0.40 |
FAM110A |
family with sequence similarity 110, member A |
1486 |
0.47 |
| chr2_74433191_74433522 | 0.40 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
7613 |
0.14 |
| chr1_154984608_154984759 | 0.40 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
2241 |
0.12 |
| chr8_134084504_134084857 | 0.39 |
SLA |
Src-like-adaptor |
12077 |
0.24 |
| chr22_50631230_50631436 | 0.39 |
TRABD |
TraB domain containing |
110 |
0.82 |
| chr14_22987545_22987798 | 0.39 |
TRAJ15 |
T cell receptor alpha joining 15 |
10909 |
0.1 |
| chr17_56410108_56410593 | 0.38 |
MIR142 |
microRNA 142 |
481 |
0.66 |
| chr1_40849833_40850457 | 0.38 |
SMAP2 |
small ArfGAP2 |
9825 |
0.18 |
| chr11_95420136_95420287 | 0.37 |
FAM76B |
family with sequence similarity 76, member B |
99758 |
0.08 |
| chr13_114428270_114428568 | 0.36 |
TMEM255B |
transmembrane protein 255B |
33797 |
0.18 |
| chr2_127422219_127422370 | 0.36 |
GYPC |
glycophorin C (Gerbich blood group) |
8534 |
0.27 |
| chr2_231588412_231588687 | 0.36 |
CAB39 |
calcium binding protein 39 |
10286 |
0.21 |
| chr1_39680841_39681106 | 0.35 |
RP11-416A14.1 |
|
8789 |
0.18 |
| chrY_14775150_14775569 | 0.35 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
37801 |
0.23 |
| chr16_28305279_28305430 | 0.35 |
SBK1 |
SH3 domain binding kinase 1 |
1514 |
0.42 |
| chr17_40439817_40440043 | 0.34 |
STAT5A |
signal transducer and activator of transcription 5A |
45 |
0.96 |
| chr22_47070453_47070681 | 0.34 |
GRAMD4 |
GRAM domain containing 4 |
62 |
0.98 |
| chr7_142495656_142495944 | 0.34 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
14669 |
0.17 |
| chr12_55377683_55377834 | 0.33 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
698 |
0.74 |
| chr17_56413846_56414087 | 0.33 |
ENSG00000264399 |
. |
583 |
0.4 |
| chr1_1830918_1831069 | 0.33 |
RP1-140A9.1 |
|
8083 |
0.13 |
| chr18_21581201_21581352 | 0.33 |
TTC39C |
tetratricopeptide repeat domain 39C |
8539 |
0.18 |
| chr16_3627016_3627406 | 0.33 |
NLRC3 |
NLR family, CARD domain containing 3 |
181 |
0.93 |
| chr12_15113209_15113383 | 0.33 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
904 |
0.55 |
| chr2_86088659_86088948 | 0.32 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
5974 |
0.2 |
| chr15_65185686_65185837 | 0.32 |
ENSG00000264929 |
. |
6677 |
0.16 |
| chrX_118825837_118826063 | 0.32 |
SEPT6 |
septin 6 |
842 |
0.6 |
| chr2_70321532_70321889 | 0.31 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
5732 |
0.17 |
| chr12_122230087_122230336 | 0.31 |
RHOF |
ras homolog family member F (in filopodia) |
1055 |
0.48 |
| chr9_132650530_132650755 | 0.30 |
FNBP1 |
formin binding protein 1 |
30947 |
0.13 |
| chr6_53209040_53209191 | 0.30 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
4472 |
0.24 |
| chr13_31309265_31309542 | 0.30 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
242 |
0.96 |
| chr8_22976794_22976945 | 0.30 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
16435 |
0.13 |
| chr11_3969184_3969451 | 0.29 |
STIM1 |
stromal interaction molecule 1 |
744 |
0.65 |
| chr19_48760988_48761397 | 0.29 |
CARD8 |
caspase recruitment domain family, member 8 |
1989 |
0.2 |
| chr1_208040719_208040870 | 0.29 |
CD34 |
CD34 molecule |
23043 |
0.23 |
| chr4_109082592_109082835 | 0.29 |
LEF1 |
lymphoid enhancer-binding factor 1 |
4744 |
0.25 |
| chr11_18267936_18268087 | 0.29 |
SAA2 |
serum amyloid A2 |
1555 |
0.21 |
| chr11_118210860_118211247 | 0.28 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
599 |
0.61 |
| chr6_10529624_10529892 | 0.28 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
1169 |
0.49 |
| chr1_74475780_74475968 | 0.28 |
LRRIQ3 |
leucine-rich repeats and IQ motif containing 3 |
64600 |
0.13 |
| chr19_10842481_10842632 | 0.28 |
DNM2 |
dynamin 2 |
13319 |
0.11 |
| chr17_73840046_73840332 | 0.28 |
UNC13D |
unc-13 homolog D (C. elegans) |
226 |
0.84 |
| chr18_2981073_2981432 | 0.28 |
LPIN2 |
lipin 2 |
1619 |
0.33 |
| chr17_45811642_45811793 | 0.28 |
TBX21 |
T-box 21 |
1107 |
0.44 |
| chrX_48793999_48794379 | 0.28 |
PIM2 |
pim-2 oncogene |
17888 |
0.08 |
| chr11_104915569_104916017 | 0.28 |
CARD16 |
caspase recruitment domain family, member 16 |
241 |
0.92 |
| chr10_15296464_15296906 | 0.28 |
RP11-25G10.2 |
|
17194 |
0.24 |
| chr2_162929797_162930173 | 0.27 |
AC008063.2 |
|
219 |
0.89 |
| chr11_67186305_67186456 | 0.27 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
2272 |
0.11 |
| chr12_51716594_51716774 | 0.27 |
BIN2 |
bridging integrator 2 |
1215 |
0.42 |
| chr8_27236010_27236326 | 0.27 |
PTK2B |
protein tyrosine kinase 2 beta |
2000 |
0.39 |
| chr1_117306495_117306758 | 0.27 |
CD2 |
CD2 molecule |
9537 |
0.22 |
| chrY_7140936_7141087 | 0.27 |
PRKY |
protein kinase, Y-linked, pseudogene |
1325 |
0.53 |
| chr20_56202934_56203085 | 0.27 |
ZBP1 |
Z-DNA binding protein 1 |
7377 |
0.24 |
| chr14_91854852_91855042 | 0.27 |
CCDC88C |
coiled-coil domain containing 88C |
28743 |
0.19 |
| chr11_76794049_76794313 | 0.27 |
CAPN5 |
calpain 5 |
16141 |
0.16 |
| chr5_176859853_176860004 | 0.27 |
GRK6 |
G protein-coupled receptor kinase 6 |
6073 |
0.1 |
| chr13_41187779_41187930 | 0.26 |
FOXO1 |
forkhead box O1 |
52880 |
0.14 |
| chr5_79487674_79487825 | 0.26 |
ENSG00000239159 |
. |
47659 |
0.12 |
| chr12_25204773_25204947 | 0.26 |
LRMP |
lymphoid-restricted membrane protein |
345 |
0.89 |
| chr8_144107989_144108192 | 0.26 |
LY6E |
lymphocyte antigen 6 complex, locus E |
8116 |
0.17 |
| chr16_85979656_85979807 | 0.26 |
IRF8 |
interferon regulatory factor 8 |
31812 |
0.2 |
| chr5_95171463_95171720 | 0.26 |
GLRX |
glutaredoxin (thioltransferase) |
12882 |
0.13 |
| chr5_159713959_159714110 | 0.26 |
ENSG00000243654 |
. |
4761 |
0.2 |
| chrX_105621857_105622008 | 0.25 |
ENSG00000202407 |
. |
47866 |
0.19 |
| chr7_26704596_26704975 | 0.25 |
C7orf71 |
chromosome 7 open reading frame 71 |
27295 |
0.23 |
| chr9_123688442_123688941 | 0.25 |
TRAF1 |
TNF receptor-associated factor 1 |
2356 |
0.33 |
| chr6_29911310_29911740 | 0.25 |
HLA-A |
major histocompatibility complex, class I, A |
1199 |
0.47 |
| chr16_84641211_84641362 | 0.25 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
10379 |
0.17 |
| chr15_64984890_64985206 | 0.25 |
AC100830.4 |
|
2192 |
0.18 |
| chr6_117870110_117870478 | 0.25 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
53233 |
0.12 |
| chr16_85768972_85769153 | 0.24 |
ENSG00000222190 |
. |
6244 |
0.13 |
| chr8_8777943_8778119 | 0.24 |
ENSG00000200713 |
. |
24708 |
0.16 |
| chr17_2717574_2718059 | 0.24 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
18040 |
0.17 |
| chr5_133458466_133458617 | 0.24 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
768 |
0.69 |
| chr17_76123369_76123520 | 0.24 |
TMC6 |
transmembrane channel-like 6 |
343 |
0.8 |
| chr13_100072035_100072186 | 0.24 |
ENSG00000266207 |
. |
33613 |
0.16 |
| chr10_45874852_45875003 | 0.24 |
ALOX5 |
arachidonate 5-lipoxygenase |
5252 |
0.27 |
| chr2_238166258_238166409 | 0.24 |
AC112715.2 |
Uncharacterized protein |
599 |
0.83 |
| chr2_198167316_198167467 | 0.24 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
148 |
0.95 |
| chr13_46745397_46745548 | 0.24 |
ENSG00000240767 |
. |
1589 |
0.32 |
| chr11_104970539_104970718 | 0.24 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
1530 |
0.24 |
| chr14_22997122_22997273 | 0.24 |
TRAJ15 |
T cell receptor alpha joining 15 |
1383 |
0.28 |
| chr12_50561431_50561798 | 0.24 |
CERS5 |
ceramide synthase 5 |
409 |
0.77 |
| chr14_52818673_52818824 | 0.23 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
37635 |
0.19 |
| chr11_104940583_104940734 | 0.23 |
CARD16 |
caspase recruitment domain family, member 16 |
24555 |
0.15 |
| chr7_26332543_26332766 | 0.23 |
SNX10 |
sorting nexin 10 |
20 |
0.98 |
| chr19_14496223_14496374 | 0.23 |
CD97 |
CD97 molecule |
4042 |
0.17 |
| chr7_5183321_5183472 | 0.23 |
ZNF890P |
zinc finger protein 890, pseudogene |
15970 |
0.17 |
| chr7_37393476_37393702 | 0.23 |
ELMO1 |
engulfment and cell motility 1 |
317 |
0.89 |
| chr17_79259389_79259896 | 0.23 |
SLC38A10 |
solute carrier family 38, member 10 |
3578 |
0.16 |
| chr3_53205926_53206171 | 0.23 |
PRKCD |
protein kinase C, delta |
6903 |
0.18 |
| chr2_202125315_202125762 | 0.23 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
315 |
0.9 |
| chr5_176857238_176857389 | 0.23 |
GRK6 |
G protein-coupled receptor kinase 6 |
3458 |
0.12 |
| chr22_50752772_50753385 | 0.22 |
XX-C283C717.1 |
|
18 |
0.95 |
| chr1_234749993_234750144 | 0.22 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
4797 |
0.21 |
| chr19_10714199_10714545 | 0.22 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
1186 |
0.31 |
| chr12_9912992_9913534 | 0.22 |
CD69 |
CD69 molecule |
234 |
0.92 |
| chr22_47131590_47131741 | 0.22 |
CERK |
ceramide kinase |
2439 |
0.28 |
| chr17_47816551_47816978 | 0.22 |
FAM117A |
family with sequence similarity 117, member A |
14875 |
0.14 |
| chr7_38313129_38313494 | 0.22 |
STARD3NL |
STARD3 N-terminal like |
95314 |
0.09 |
| chr1_236101843_236102121 | 0.21 |
ENSG00000206803 |
. |
23267 |
0.18 |
| chr14_94437286_94437966 | 0.21 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2803 |
0.22 |
| chr12_54718303_54718481 | 0.21 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
519 |
0.57 |
| chr6_30460368_30460519 | 0.21 |
HLA-E |
major histocompatibility complex, class I, E |
3199 |
0.18 |
| chr19_18388821_18388972 | 0.21 |
JUND |
jun D proto-oncogene |
2843 |
0.13 |
| chr18_77163060_77163265 | 0.21 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
2770 |
0.33 |
| chr19_49838375_49839003 | 0.21 |
CD37 |
CD37 molecule |
5 |
0.95 |
| chr8_23075544_23076103 | 0.21 |
ENSG00000246582 |
. |
6161 |
0.12 |
| chr14_24640773_24641022 | 0.21 |
REC8 |
REC8 meiotic recombination protein |
165 |
0.84 |
| chr1_9486725_9486876 | 0.21 |
ENSG00000252956 |
. |
11037 |
0.24 |
| chr12_12223823_12224258 | 0.21 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
167 |
0.97 |
| chr17_72459969_72460120 | 0.21 |
CD300A |
CD300a molecule |
2511 |
0.22 |
| chr5_133467374_133467525 | 0.21 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
8140 |
0.21 |
| chr10_107262399_107262550 | 0.21 |
ENSG00000207068 |
. |
31943 |
0.26 |
| chr17_33672914_33673171 | 0.21 |
SLFN11 |
schlafen family member 11 |
7108 |
0.2 |
| chr1_46153318_46153689 | 0.21 |
TMEM69 |
transmembrane protein 69 |
617 |
0.57 |
| chr1_111162215_111162656 | 0.20 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
11661 |
0.19 |
| chr1_160709134_160709343 | 0.20 |
SLAMF7 |
SLAM family member 7 |
144 |
0.95 |
| chr21_46347092_46347406 | 0.20 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
1384 |
0.25 |
| chr21_40176782_40176933 | 0.20 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
374 |
0.92 |
| chr20_49548700_49548918 | 0.20 |
ADNP |
activity-dependent neuroprotector homeobox |
851 |
0.42 |
| chr12_54610003_54610573 | 0.20 |
ENSG00000265371 |
. |
14972 |
0.1 |
| chr16_30196259_30196584 | 0.20 |
RP11-455F5.5 |
|
143 |
0.71 |
| chr19_57100792_57100943 | 0.20 |
ZNF71 |
zinc finger protein 71 |
5765 |
0.14 |
| chr2_149285721_149285937 | 0.20 |
MBD5 |
methyl-CpG binding domain protein 5 |
59535 |
0.15 |
| chr5_156693510_156694312 | 0.20 |
CTC-248O19.1 |
|
276 |
0.76 |
| chr17_41445494_41445820 | 0.20 |
ENSG00000188825 |
. |
19128 |
0.11 |
| chr16_1014386_1014626 | 0.20 |
LMF1 |
lipase maturation factor 1 |
384 |
0.78 |
| chr10_75716254_75716492 | 0.20 |
C10orf55 |
chromosome 10 open reading frame 55 |
33838 |
0.11 |
| chr2_26635298_26635481 | 0.19 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
10605 |
0.21 |
| chr11_64631996_64632371 | 0.19 |
EHD1 |
EH-domain containing 1 |
10955 |
0.1 |
| chrX_152200996_152201209 | 0.19 |
PNMA3 |
paraneoplastic Ma antigen 3 |
23664 |
0.12 |
| chr1_34542680_34542831 | 0.19 |
ENSG00000201148 |
. |
35795 |
0.19 |
| chr9_138999016_138999167 | 0.19 |
C9orf69 |
chromosome 9 open reading frame 69 |
11029 |
0.19 |
| chr19_17516551_17516849 | 0.19 |
MVB12A |
multivesicular body subunit 12A |
150 |
0.6 |
| chr9_134273565_134273749 | 0.19 |
PRRC2B |
proline-rich coiled-coil 2B |
4177 |
0.25 |
| chr1_160489868_160490211 | 0.19 |
SLAMF6 |
SLAM family member 6 |
2993 |
0.18 |
| chr6_13274566_13274804 | 0.19 |
RP1-257A7.4 |
|
394 |
0.82 |
| chr12_9800952_9801457 | 0.19 |
RP11-705C15.4 |
|
599 |
0.62 |
| chr8_82022969_82023806 | 0.19 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
916 |
0.72 |
| chr1_29253197_29253478 | 0.19 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12246 |
0.18 |
| chr20_61407912_61408071 | 0.18 |
LINC00659 |
long intergenic non-protein coding RNA 659 |
1227 |
0.34 |
| chr3_71115551_71115702 | 0.18 |
FOXP1 |
forkhead box P1 |
708 |
0.81 |
| chr6_37141288_37141439 | 0.18 |
PIM1 |
pim-1 oncogene |
3384 |
0.23 |
| chr14_90866152_90866669 | 0.18 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
1804 |
0.32 |
| chr19_54875923_54876565 | 0.18 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
170 |
0.91 |
| chrX_123481870_123482040 | 0.18 |
SH2D1A |
SH2 domain containing 1A |
1516 |
0.57 |
| chr11_14995196_14995439 | 0.18 |
CALCA |
calcitonin-related polypeptide alpha |
1417 |
0.53 |
| chr17_76642017_76642260 | 0.18 |
ENSG00000252391 |
. |
50741 |
0.12 |
| chr3_194871620_194871771 | 0.18 |
XXYLT1-AS2 |
XXYLT1 antisense RNA 2 |
3075 |
0.19 |
| chr13_30510013_30510639 | 0.18 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
9538 |
0.3 |
| chr19_16477276_16478284 | 0.18 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
5016 |
0.17 |
| chr9_130731987_130732222 | 0.18 |
FAM102A |
family with sequence similarity 102, member A |
10688 |
0.11 |
| chr8_103898951_103899102 | 0.18 |
KB-1507C5.3 |
|
5078 |
0.18 |
| chr20_33680803_33681037 | 0.18 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
246 |
0.9 |
| chr1_111745814_111746254 | 0.18 |
DENND2D |
DENN/MADD domain containing 2D |
997 |
0.41 |
| chr6_144665292_144666004 | 0.18 |
UTRN |
utrophin |
411 |
0.88 |
| chr17_38018344_38018495 | 0.18 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1960 |
0.26 |
| chr17_18161547_18161698 | 0.18 |
FLII |
flightless I homolog (Drosophila) |
248 |
0.85 |
| chr15_38853827_38853978 | 0.18 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
1422 |
0.42 |
| chr17_36859477_36859845 | 0.18 |
ENSG00000265930 |
. |
1077 |
0.24 |
| chr14_25142934_25143265 | 0.18 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
39626 |
0.14 |
| chr17_33567122_33567465 | 0.18 |
RP11-799D4.4 |
|
2293 |
0.22 |
| chr1_117343275_117343583 | 0.17 |
CD2 |
CD2 molecule |
46340 |
0.14 |
| chr10_14606149_14606418 | 0.17 |
FAM107B |
family with sequence similarity 107, member B |
7746 |
0.26 |
| chr5_180633345_180633855 | 0.17 |
CTC-338M12.1 |
|
480 |
0.54 |
| chr20_35203812_35203967 | 0.17 |
TGIF2 |
TGFB-induced factor homeobox 2 |
814 |
0.36 |
| chr14_22963495_22963646 | 0.17 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
7399 |
0.11 |
| chr16_81503920_81504132 | 0.17 |
CMIP |
c-Maf inducing protein |
24928 |
0.22 |
| chr7_50357605_50357756 | 0.17 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
9362 |
0.29 |
| chr2_45169165_45169332 | 0.17 |
SIX3-AS1 |
SIX3 antisense RNA 1 |
236 |
0.62 |
| chr1_29254208_29254684 | 0.17 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
13355 |
0.18 |
| chr17_44269690_44270135 | 0.17 |
KANSL1 |
KAT8 regulatory NSL complex subunit 1 |
254 |
0.85 |
| chr3_69247967_69248118 | 0.17 |
FRMD4B |
FERM domain containing 4B |
379 |
0.9 |
| chr7_99765104_99765382 | 0.17 |
GAL3ST4 |
galactose-3-O-sulfotransferase 4 |
336 |
0.69 |
| chr20_47382876_47383027 | 0.17 |
ENSG00000251876 |
. |
26966 |
0.22 |
| chr8_33371053_33371277 | 0.17 |
TTI2 |
TELO2 interacting protein 2 |
46 |
0.66 |
| chr4_143741757_143741908 | 0.17 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
25611 |
0.28 |
| chr2_143934931_143935154 | 0.17 |
RP11-190J23.1 |
|
5301 |
0.3 |
| chr8_134079225_134079805 | 0.17 |
SLA |
Src-like-adaptor |
6912 |
0.26 |
| chr19_6802455_6802606 | 0.17 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
18401 |
0.13 |
| chr6_168501309_168501460 | 0.17 |
FRMD1 |
FERM domain containing 1 |
19147 |
0.25 |
| chr14_22963855_22964006 | 0.17 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
7759 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.3 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.8 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.4 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.1 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.4 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.2 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) NK T cell activation(GO:0051132) |
| 0.0 | 0.1 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.2 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.0 | GO:0048305 | immunoglobulin secretion(GO:0048305) regulation of immunoglobulin secretion(GO:0051023) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0019054 | modulation by virus of host process(GO:0019054) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 1.2 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.0 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:0002874 | chronic inflammatory response to antigenic stimulus(GO:0002439) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.8 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |