Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
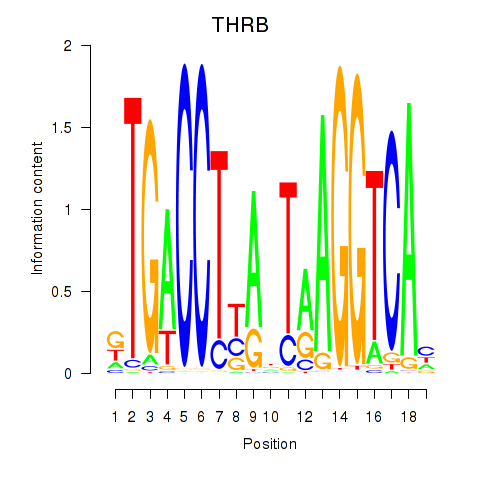
Results for THRB
Z-value: 0.37
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | thyroid hormone receptor beta |
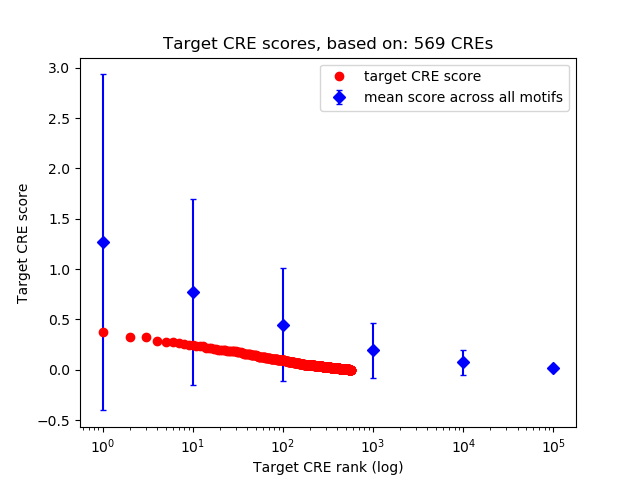
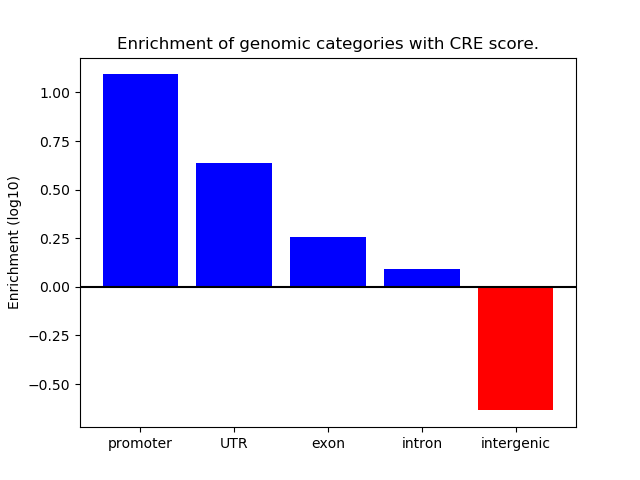
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_24101464_24101651 | THRB | 105529 | 0.065211 | -0.85 | 4.1e-03 | Click! |
| chr3_24283504_24283655 | THRB | 76493 | 0.116760 | -0.83 | 6.0e-03 | Click! |
| chr3_24218786_24218937 | THRB | 11775 | 0.299154 | -0.82 | 6.5e-03 | Click! |
| chr3_24263536_24263687 | THRB | 56525 | 0.162538 | -0.80 | 1.0e-02 | Click! |
| chr3_24548896_24549047 | THRB | 12198 | 0.177837 | -0.78 | 1.3e-02 | Click! |
Activity of the THRB motif across conditions
Conditions sorted by the z-value of the THRB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
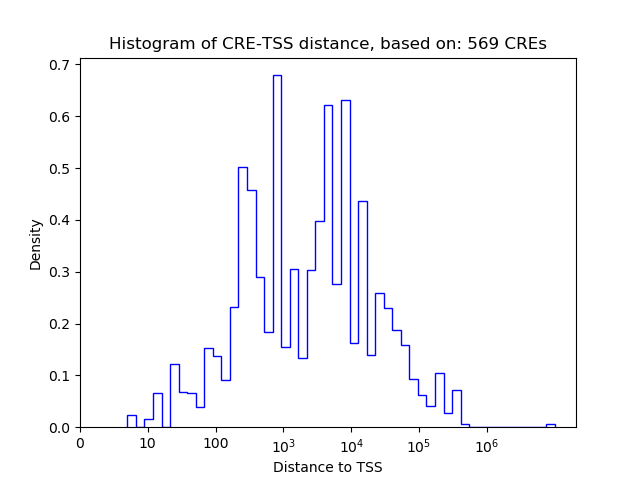
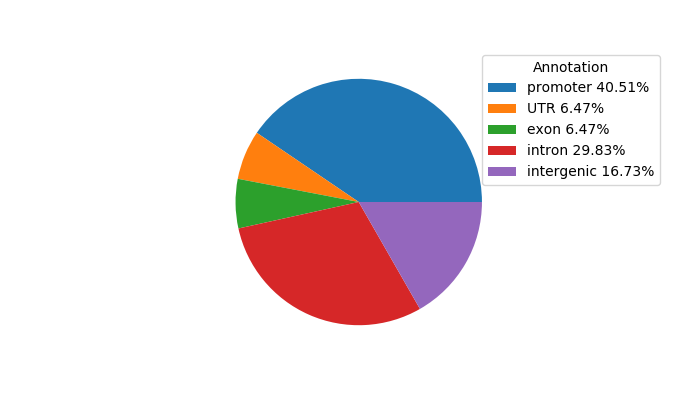
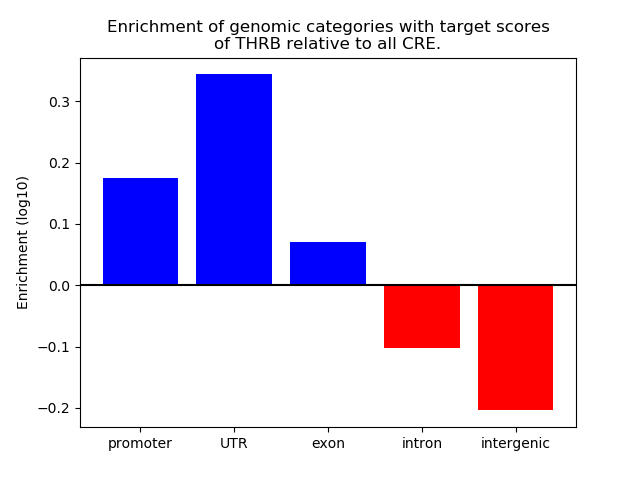
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_118094529_118095128 | 0.37 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
26 |
0.97 |
| chr14_22963855_22964006 | 0.33 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
7759 |
0.1 |
| chr17_73840046_73840332 | 0.33 |
UNC13D |
unc-13 homolog D (C. elegans) |
226 |
0.84 |
| chr8_128987632_128988045 | 0.29 |
ENSG00000221771 |
. |
14959 |
0.16 |
| chr2_106392629_106392859 | 0.28 |
NCK2 |
NCK adaptor protein 2 |
30556 |
0.23 |
| chr17_72749074_72749225 | 0.28 |
MIR3615 |
microRNA 3615 |
4098 |
0.11 |
| chr13_105791565_105791981 | 0.27 |
DAOA |
D-amino acid oxidase activator |
326819 |
0.01 |
| chr13_34184631_34184800 | 0.25 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
207471 |
0.02 |
| chr18_77283391_77284187 | 0.25 |
AC018445.1 |
Uncharacterized protein |
7732 |
0.29 |
| chr10_114135452_114136486 | 0.24 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
12 |
0.98 |
| chr1_241715191_241715459 | 0.24 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
16610 |
0.2 |
| chr1_36786583_36786988 | 0.23 |
RP11-268J15.5 |
|
2550 |
0.19 |
| chr11_3858945_3859096 | 0.23 |
RHOG |
ras homolog family member G |
77 |
0.94 |
| chr15_31623203_31623354 | 0.21 |
KLF13 |
Kruppel-like factor 13 |
4220 |
0.35 |
| chr11_73499157_73499522 | 0.21 |
MRPL48 |
mitochondrial ribosomal protein L48 |
364 |
0.84 |
| chr1_203277833_203278085 | 0.21 |
BTG2 |
BTG family, member 2 |
3295 |
0.22 |
| chr2_10472194_10472345 | 0.20 |
HPCAL1 |
hippocalcin-like 1 |
28443 |
0.15 |
| chr14_102281143_102281546 | 0.20 |
CTD-2017C7.2 |
|
4686 |
0.17 |
| chrX_11781800_11781951 | 0.20 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
4128 |
0.35 |
| chr1_151798177_151798381 | 0.20 |
RORC |
RAR-related orphan receptor C |
288 |
0.78 |
| chr12_117536177_117536841 | 0.20 |
TESC |
tescalcin |
742 |
0.76 |
| chr22_30819167_30819485 | 0.20 |
RNF215 |
ring finger protein 215 |
1566 |
0.14 |
| chr8_27237778_27238009 | 0.19 |
PTK2B |
protein tyrosine kinase 2 beta |
275 |
0.93 |
| chr12_26351665_26351816 | 0.19 |
SSPN |
sarcospan |
3134 |
0.29 |
| chr19_1068246_1068401 | 0.19 |
HMHA1 |
histocompatibility (minor) HA-1 |
826 |
0.4 |
| chr8_74657868_74658768 | 0.19 |
STAU2 |
staufen double-stranded RNA binding protein 2 |
757 |
0.75 |
| chr3_187803854_187804005 | 0.19 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
67143 |
0.12 |
| chr12_53719733_53719917 | 0.19 |
AAAS |
achalasia, adrenocortical insufficiency, alacrimia |
1283 |
0.29 |
| chr17_38490189_38490340 | 0.19 |
RARA |
retinoic acid receptor, alpha |
7376 |
0.11 |
| chr3_50606285_50606482 | 0.19 |
HEMK1 |
HemK methyltransferase family member 1 |
200 |
0.89 |
| chr3_41159090_41159342 | 0.18 |
CTNNB1 |
catenin (cadherin-associated protein), beta 1, 88kDa |
77112 |
0.12 |
| chr5_148726870_148727055 | 0.18 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
1897 |
0.22 |
| chr12_7063274_7063561 | 0.17 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
2879 |
0.08 |
| chr1_78546388_78546598 | 0.17 |
ENSG00000251958 |
. |
8274 |
0.17 |
| chr9_139267510_139268079 | 0.17 |
CARD9 |
caspase recruitment domain family, member 9 |
339 |
0.79 |
| chr8_28601710_28601861 | 0.16 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
14108 |
0.17 |
| chr6_42332393_42332544 | 0.16 |
ENSG00000221252 |
. |
47666 |
0.14 |
| chr14_98443402_98443682 | 0.16 |
C14orf64 |
chromosome 14 open reading frame 64 |
841 |
0.77 |
| chr9_130823670_130823942 | 0.16 |
RP11-379C10.1 |
|
4165 |
0.12 |
| chrX_11768810_11768961 | 0.16 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
7393 |
0.31 |
| chr5_74806864_74807381 | 0.16 |
COL4A3BP |
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
332 |
0.59 |
| chr2_177503220_177503371 | 0.15 |
ENSG00000252027 |
. |
26109 |
0.25 |
| chr14_50365431_50365582 | 0.15 |
ENSG00000251929 |
. |
3162 |
0.16 |
| chrX_40033281_40033432 | 0.15 |
BCOR |
BCL6 corepressor |
3217 |
0.38 |
| chr7_103577846_103577997 | 0.15 |
RELN |
reelin |
52042 |
0.18 |
| chr18_52985449_52985614 | 0.15 |
TCF4 |
transcription factor 4 |
3686 |
0.35 |
| chr2_30476045_30476196 | 0.15 |
LBH |
limb bud and heart development |
21074 |
0.22 |
| chr10_120924621_120924984 | 0.15 |
SFXN4 |
sideroflexin 4 |
252 |
0.89 |
| chr11_33721707_33721949 | 0.14 |
C11orf91 |
chromosome 11 open reading frame 91 |
115 |
0.96 |
| chr19_45354958_45355109 | 0.14 |
PVRL2 |
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
5250 |
0.12 |
| chr8_11725136_11725560 | 0.14 |
CTSB |
cathepsin B |
146 |
0.95 |
| chr5_175085682_175085863 | 0.14 |
HRH2 |
histamine receptor H2 |
739 |
0.7 |
| chr16_29000534_29000778 | 0.14 |
RP11-264B17.5 |
|
1443 |
0.21 |
| chr16_3124352_3124503 | 0.13 |
ENSG00000252561 |
. |
4903 |
0.07 |
| chr19_52642669_52643149 | 0.13 |
ZNF616 |
zinc finger protein 616 |
90 |
0.95 |
| chr1_169672728_169672969 | 0.13 |
SELL |
selectin L |
7991 |
0.2 |
| chr5_111071208_111071359 | 0.13 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
5197 |
0.22 |
| chr6_106967766_106968029 | 0.13 |
AIM1 |
absent in melanoma 1 |
8167 |
0.21 |
| chr6_143165477_143165628 | 0.13 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
7368 |
0.3 |
| chr22_29999046_29999267 | 0.13 |
NF2 |
neurofibromin 2 (merlin) |
389 |
0.83 |
| chr1_218833930_218834081 | 0.13 |
ENSG00000212610 |
. |
118764 |
0.06 |
| chr10_69373029_69373180 | 0.12 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
34167 |
0.23 |
| chr1_21658510_21658661 | 0.12 |
ECE1 |
endothelin converting enzyme 1 |
13412 |
0.2 |
| chr22_39843892_39844043 | 0.12 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
9382 |
0.15 |
| chr1_117112540_117113022 | 0.12 |
CD58 |
CD58 molecule |
867 |
0.56 |
| chr3_113251981_113252391 | 0.12 |
SIDT1 |
SID1 transmembrane family, member 1 |
968 |
0.54 |
| chr6_2999439_2999644 | 0.12 |
RP1-90J20.8 |
|
173 |
0.82 |
| chr11_114007906_114008308 | 0.12 |
ENSG00000221112 |
. |
50456 |
0.14 |
| chrX_71321072_71321371 | 0.12 |
RGAG4 |
retrotransposon gag domain containing 4 |
30457 |
0.14 |
| chr11_117858339_117858806 | 0.11 |
IL10RA |
interleukin 10 receptor, alpha |
1463 |
0.41 |
| chr8_93599945_93600096 | 0.11 |
ENSG00000221172 |
. |
47543 |
0.19 |
| chr12_52317139_52317323 | 0.11 |
ACVRL1 |
activin A receptor type II-like 1 |
7999 |
0.16 |
| chr13_38170462_38170613 | 0.11 |
POSTN |
periostin, osteoblast specific factor |
2326 |
0.46 |
| chr10_76727713_76728120 | 0.11 |
RP11-77G23.5 |
|
56093 |
0.12 |
| chr20_31251603_31251754 | 0.11 |
RP11-410N8.4 |
|
62239 |
0.09 |
| chr20_62367320_62367471 | 0.11 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
240 |
0.82 |
| chr15_41316618_41316840 | 0.11 |
RP11-540O11.4 |
|
32 |
0.97 |
| chr6_127590111_127590603 | 0.11 |
RNF146 |
ring finger protein 146 |
2324 |
0.36 |
| chr1_39457743_39458066 | 0.11 |
AKIRIN1 |
akirin 1 |
736 |
0.62 |
| chr1_26316819_26316970 | 0.11 |
PAFAH2 |
platelet-activating factor acetylhydrolase 2, 40kDa |
7218 |
0.1 |
| chr21_47742400_47742668 | 0.11 |
C21orf58 |
chromosome 21 open reading frame 58 |
1237 |
0.31 |
| chr7_92347863_92348047 | 0.11 |
ENSG00000206763 |
. |
16827 |
0.24 |
| chr6_32908448_32908870 | 0.11 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
120 |
0.92 |
| chr5_162864043_162864372 | 0.11 |
RP11-541P9.3 |
|
92 |
0.81 |
| chr17_3537923_3538154 | 0.11 |
SHPK |
sedoheptulokinase |
1578 |
0.2 |
| chrX_46694115_46694266 | 0.10 |
RP2 |
retinitis pigmentosa 2 (X-linked recessive) |
2185 |
0.36 |
| chr13_24024495_24024646 | 0.10 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
16729 |
0.28 |
| chrX_135848946_135849996 | 0.10 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
31 |
0.98 |
| chr19_4067333_4067959 | 0.10 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
703 |
0.54 |
| chr5_158633601_158634197 | 0.10 |
RNF145 |
ring finger protein 145 |
743 |
0.62 |
| chrY_2369174_2369325 | 0.10 |
ENSG00000251841 |
. |
283541 |
0.01 |
| chr17_39137222_39137388 | 0.10 |
KRT40 |
keratin 40 |
3255 |
0.1 |
| chr2_62445013_62445164 | 0.10 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
2435 |
0.26 |
| chr1_226069091_226069419 | 0.10 |
TMEM63A |
transmembrane protein 63A |
814 |
0.51 |
| chr1_225965575_225965972 | 0.10 |
SRP9 |
signal recognition particle 9kDa |
136 |
0.95 |
| chr21_46323297_46323448 | 0.10 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
7173 |
0.11 |
| chr13_113622985_113623455 | 0.10 |
MCF2L-AS1 |
MCF2L antisense RNA 1 |
82 |
0.79 |
| chr2_242459234_242459385 | 0.10 |
STK25 |
serine/threonine kinase 25 |
10164 |
0.13 |
| chrX_2419173_2419324 | 0.10 |
DHRSX |
dehydrogenase/reductase (SDR family) X-linked |
229 |
0.51 |
| chr1_111682881_111683246 | 0.09 |
CEPT1 |
choline/ethanolamine phosphotransferase 1 |
215 |
0.5 |
| chr17_61497297_61497448 | 0.09 |
AC015923.1 |
|
5699 |
0.14 |
| chr8_82018570_82018849 | 0.09 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
5594 |
0.32 |
| chr17_66259356_66259507 | 0.09 |
ARSG |
arylsulfatase G |
4108 |
0.19 |
| chr6_25992069_25992220 | 0.09 |
U91328.21 |
|
7251 |
0.09 |
| chr12_64375360_64375627 | 0.09 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
812 |
0.7 |
| chr14_78227118_78227433 | 0.09 |
C14orf178 |
chromosome 14 open reading frame 178 |
42 |
0.71 |
| chr1_1004234_1004415 | 0.09 |
RNF223 |
ring finger protein 223 |
5363 |
0.1 |
| chr12_52918032_52918183 | 0.09 |
KRT5 |
keratin 5 |
3636 |
0.13 |
| chr4_40201423_40201705 | 0.09 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chr9_70851276_70851465 | 0.09 |
CBWD3 |
COBW domain containing 3 |
5027 |
0.19 |
| chr17_48474498_48474752 | 0.09 |
LRRC59 |
leucine rich repeat containing 59 |
240 |
0.78 |
| chr6_88410118_88410669 | 0.09 |
AKIRIN2 |
akirin 2 |
1534 |
0.41 |
| chr10_69829246_69829489 | 0.09 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
4527 |
0.23 |
| chr16_25043620_25043907 | 0.08 |
ARHGAP17 |
Rho GTPase activating protein 17 |
16776 |
0.24 |
| chr3_150994287_150994620 | 0.08 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
1704 |
0.34 |
| chr20_4982345_4982709 | 0.08 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
374 |
0.87 |
| chr2_198363785_198364521 | 0.08 |
HSPD1 |
heat shock 60kDa protein 1 (chaperonin) |
406 |
0.5 |
| chr4_1238249_1238400 | 0.08 |
CTBP1 |
C-terminal binding protein 1 |
2613 |
0.2 |
| chr8_144821974_144822220 | 0.08 |
FAM83H |
family with sequence similarity 83, member H |
6126 |
0.08 |
| chr9_70495582_70495733 | 0.08 |
CBWD5 |
COBW domain containing 5 |
5411 |
0.29 |
| chr5_176856544_176856754 | 0.08 |
GRK6 |
G protein-coupled receptor kinase 6 |
2794 |
0.13 |
| chr9_184434_184636 | 0.08 |
CBWD1 |
COBW domain containing 1 |
5463 |
0.2 |
| chr19_6772910_6773160 | 0.08 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
66 |
0.96 |
| chr11_75062077_75062724 | 0.08 |
ARRB1 |
arrestin, beta 1 |
349 |
0.83 |
| chr9_69267993_69268163 | 0.08 |
CBWD6 |
COBW domain containing 6 |
5485 |
0.22 |
| chr15_99859278_99859429 | 0.07 |
AC022819.2 |
Uncharacterized protein |
475 |
0.82 |
| chr17_56026245_56026547 | 0.07 |
CUEDC1 |
CUE domain containing 1 |
6222 |
0.17 |
| chr1_40847512_40847991 | 0.07 |
SMAP2 |
small ArfGAP2 |
7431 |
0.18 |
| chr22_37542366_37542517 | 0.07 |
IL2RB |
interleukin 2 receptor, beta |
3589 |
0.14 |
| chr6_31796282_31796804 | 0.07 |
HSPA1B |
heat shock 70kDa protein 1B |
815 |
0.28 |
| chr19_14496223_14496374 | 0.07 |
CD97 |
CD97 molecule |
4042 |
0.17 |
| chr14_106365954_106366105 | 0.07 |
ENSG00000227108 |
. |
483 |
0.28 |
| chr7_150212054_150212338 | 0.07 |
GIMAP7 |
GTPase, IMAP family member 7 |
278 |
0.91 |
| chr17_33446349_33446789 | 0.07 |
RAD51D |
RAD51 paralog D |
4 |
0.88 |
| chr13_103452681_103453379 | 0.07 |
BIVM |
basic, immunoglobulin-like variable motif containing |
1469 |
0.24 |
| chr10_100227763_100227980 | 0.07 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
21188 |
0.19 |
| chr1_226900274_226900494 | 0.07 |
ITPKB |
inositol-trisphosphate 3-kinase B |
24775 |
0.18 |
| chrX_64195307_64195683 | 0.07 |
ZC4H2 |
zinc finger, C4H2 domain containing |
833 |
0.78 |
| chr15_85524205_85524489 | 0.07 |
PDE8A |
phosphodiesterase 8A |
603 |
0.74 |
| chr21_46337373_46337524 | 0.07 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
3250 |
0.13 |
| chr8_144650355_144651121 | 0.07 |
MROH6 |
maestro heat-like repeat family member 6 |
286 |
0.75 |
| chr12_122238096_122238378 | 0.07 |
RHOF |
ras homolog family member F (in filopodia) |
346 |
0.85 |
| chr7_150940826_150940977 | 0.07 |
RP4-548D19.3 |
|
3416 |
0.12 |
| chr1_26872331_26872899 | 0.07 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
272 |
0.88 |
| chr6_30312997_30313362 | 0.07 |
RPP21 |
ribonuclease P/MRP 21kDa subunit |
233 |
0.9 |
| chr5_142225814_142225972 | 0.07 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
22582 |
0.24 |
| chr6_12957382_12958434 | 0.06 |
PHACTR1 |
phosphatase and actin regulator 1 |
306 |
0.95 |
| chr13_22057996_22058198 | 0.06 |
ZDHHC20 |
zinc finger, DHHC-type containing 20 |
24588 |
0.18 |
| chr13_20693229_20693593 | 0.06 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
41777 |
0.15 |
| chr20_30358146_30358468 | 0.06 |
TPX2 |
TPX2, microtubule-associated |
31233 |
0.11 |
| chr11_66059397_66059767 | 0.06 |
TMEM151A |
transmembrane protein 151A |
241 |
0.77 |
| chr1_20834118_20834607 | 0.06 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
292 |
0.9 |
| chr1_113004362_113004513 | 0.06 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
4726 |
0.24 |
| chr16_87840687_87840855 | 0.06 |
RP4-536B24.2 |
|
29367 |
0.14 |
| chr5_149340361_149340979 | 0.06 |
SLC26A2 |
solute carrier family 26 (anion exchanger), member 2 |
322 |
0.85 |
| chr22_37963047_37963198 | 0.06 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
2709 |
0.17 |
| chr20_52702899_52703050 | 0.06 |
BCAS1 |
breast carcinoma amplified sequence 1 |
15670 |
0.21 |
| chr6_74098437_74099264 | 0.06 |
ENSG00000238464 |
. |
4886 |
0.11 |
| chr2_198160747_198160898 | 0.06 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
6421 |
0.18 |
| chr6_82461200_82461724 | 0.06 |
FAM46A |
family with sequence similarity 46, member A |
8 |
0.98 |
| chr1_27995858_27996192 | 0.06 |
RP11-288L9.4 |
|
46 |
0.95 |
| chr15_102286074_102286500 | 0.06 |
RP11-89K11.1 |
Uncharacterized protein |
374 |
0.81 |
| chr11_112096740_112097015 | 0.06 |
PTS |
6-pyruvoyltetrahydropterin synthase |
211 |
0.89 |
| chr12_44151933_44152435 | 0.06 |
PUS7L |
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
377 |
0.61 |
| chr2_112249108_112249538 | 0.06 |
ENSG00000266139 |
. |
170655 |
0.04 |
| chr19_41305465_41306382 | 0.06 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
183 |
0.91 |
| chr11_307271_307422 | 0.05 |
IFITM2 |
interferon induced transmembrane protein 2 |
285 |
0.75 |
| chr11_313467_313665 | 0.05 |
IFITM1 |
interferon induced transmembrane protein 1 |
39 |
0.92 |
| chr19_6775082_6775336 | 0.05 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
2240 |
0.2 |
| chr7_23530205_23530826 | 0.05 |
ENSG00000200847 |
. |
997 |
0.52 |
| chr12_96591293_96591456 | 0.05 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
2982 |
0.28 |
| chr2_87758297_87758504 | 0.05 |
RP11-1399P15.1 |
|
19153 |
0.27 |
| chr10_17217419_17217634 | 0.05 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
867 |
0.61 |
| chr20_43997209_43997441 | 0.05 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
5231 |
0.12 |
| chr16_11835895_11836525 | 0.05 |
RP11-490O6.2 |
|
444 |
0.48 |
| chr19_42381868_42382033 | 0.05 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
575 |
0.62 |
| chr22_39268517_39268976 | 0.05 |
CBX6 |
chromobox homolog 6 |
427 |
0.79 |
| chr6_147198107_147198258 | 0.05 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
26041 |
0.25 |
| chr17_27716871_27717334 | 0.05 |
TAOK1 |
TAO kinase 1 |
380 |
0.47 |
| chr16_88768760_88768986 | 0.05 |
RNF166 |
ring finger protein 166 |
1152 |
0.23 |
| chr9_132251498_132251652 | 0.05 |
ENSG00000264298 |
. |
10740 |
0.21 |
| chr11_85565230_85565985 | 0.05 |
AP000974.1 |
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
379 |
0.6 |
| chr8_66843424_66843611 | 0.05 |
PDE7A |
phosphodiesterase 7A |
89330 |
0.09 |
| chr10_131652031_131652182 | 0.05 |
ENSG00000266676 |
. |
10468 |
0.24 |
| chr19_33726515_33726666 | 0.05 |
SLC7A10 |
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
9834 |
0.13 |
| chr17_4457464_4457615 | 0.05 |
MYBBP1A |
MYB binding protein (P160) 1a |
668 |
0.57 |
| chr14_56381006_56381157 | 0.05 |
ENSG00000212522 |
. |
154080 |
0.04 |
| chr10_126385128_126385308 | 0.05 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
6976 |
0.22 |
| chr10_71318575_71318726 | 0.05 |
NEUROG3 |
neurogenin 3 |
14344 |
0.18 |
| chr17_46872784_46872935 | 0.05 |
TTLL6 |
tubulin tyrosine ligase-like family, member 6 |
1163 |
0.37 |
| chr14_105948104_105948408 | 0.05 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
4398 |
0.12 |
| chr2_134880898_134881049 | 0.05 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
3220 |
0.29 |
| chr11_62621592_62621969 | 0.05 |
ENSG00000255717 |
. |
23 |
0.93 |
| chr10_92922169_92922499 | 0.05 |
PCGF5 |
polycomb group ring finger 5 |
57574 |
0.16 |
| chr1_201709300_201709781 | 0.05 |
NAV1 |
neuron navigator 1 |
544 |
0.66 |
| chr3_155462002_155462501 | 0.05 |
PLCH1 |
phospholipase C, eta 1 |
605 |
0.51 |
| chr11_61748704_61748855 | 0.05 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
13326 |
0.13 |
| chr6_31784100_31784303 | 0.05 |
HSPA1L |
heat shock 70kDa protein 1-like |
764 |
0.25 |
| chr12_57022963_57023540 | 0.05 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
744 |
0.54 |
| chr21_15755875_15756452 | 0.05 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
655 |
0.8 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.3 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.2 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:1904377 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |