Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
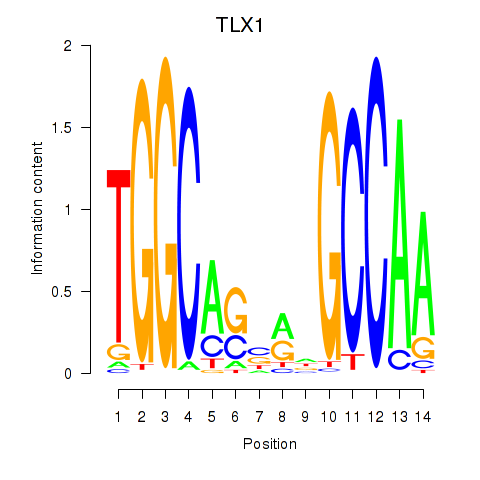
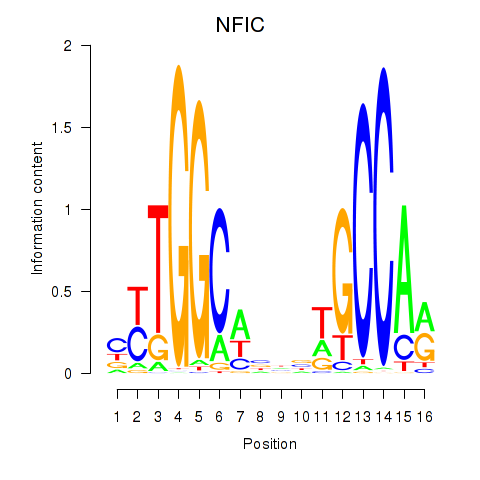
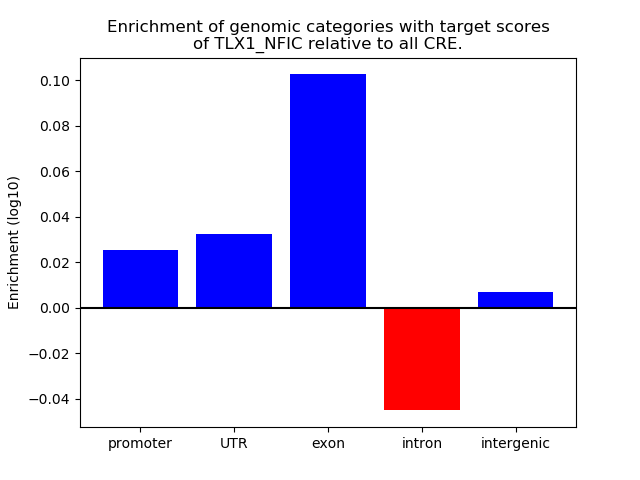
Results for TLX1_NFIC
Z-value: 1.68
Transcription factors associated with TLX1_NFIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX1
|
ENSG00000107807.8 | T cell leukemia homeobox 1 |
|
NFIC
|
ENSG00000141905.13 | nuclear factor I C |
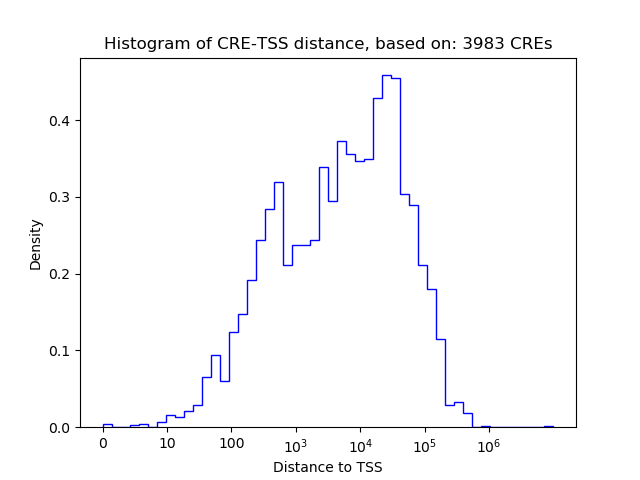
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_3346811_3347036 | NFIC | 12638 | 0.174757 | 0.85 | 3.4e-03 | Click! |
| chr19_3358174_3358371 | NFIC | 1289 | 0.448410 | -0.81 | 7.5e-03 | Click! |
| chr19_3363597_3363748 | NFIC | 2875 | 0.254994 | 0.78 | 1.3e-02 | Click! |
| chr19_3397384_3397535 | NFIC | 30875 | 0.133097 | 0.77 | 1.6e-02 | Click! |
| chr19_3368308_3368459 | NFIC | 1799 | 0.345534 | 0.70 | 3.6e-02 | Click! |
| chr10_102894531_102894816 | TLX1 | 3612 | 0.145047 | 0.51 | 1.6e-01 | Click! |
| chr10_102894012_102894163 | TLX1 | 3026 | 0.156812 | 0.39 | 3.1e-01 | Click! |
| chr10_102895075_102895226 | TLX1 | 4089 | 0.138354 | -0.36 | 3.4e-01 | Click! |
| chr10_102894867_102895065 | TLX1 | 3905 | 0.140720 | 0.20 | 6.0e-01 | Click! |
| chr10_102891889_102892040 | TLX1 | 903 | 0.360024 | 0.12 | 7.7e-01 | Click! |
Activity of the TLX1_NFIC motif across conditions
Conditions sorted by the z-value of the TLX1_NFIC motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
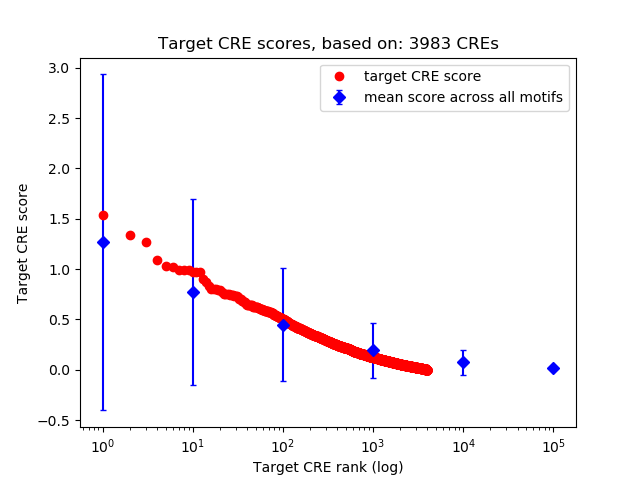
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_16389894_16390114 | 1.54 |
FAM211A |
family with sequence similarity 211, member A |
5328 |
0.18 |
| chr4_158143046_158143598 | 1.33 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
504 |
0.84 |
| chr16_4366907_4367444 | 1.27 |
GLIS2 |
GLIS family zinc finger 2 |
2413 |
0.21 |
| chr9_140506939_140507169 | 1.09 |
C9orf37 |
chromosome 9 open reading frame 37 |
6211 |
0.12 |
| chr14_75421223_75421487 | 1.03 |
PGF |
placental growth factor |
590 |
0.68 |
| chr20_45989388_45989864 | 1.02 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
4059 |
0.17 |
| chr5_115697275_115697566 | 0.99 |
CTB-118N6.3 |
|
85823 |
0.09 |
| chr8_55294582_55295235 | 0.99 |
ENSG00000244107 |
. |
38120 |
0.17 |
| chr8_65488422_65488732 | 0.99 |
RP11-21C4.1 |
|
1243 |
0.52 |
| chr1_212810031_212810274 | 0.98 |
RP11-338C15.5 |
|
10039 |
0.14 |
| chr9_109459969_109460612 | 0.97 |
ENSG00000200131 |
. |
18032 |
0.28 |
| chr19_1154555_1155129 | 0.97 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr9_25678314_25678778 | 0.90 |
TUSC1 |
tumor suppressor candidate 1 |
310 |
0.95 |
| chr10_17390339_17390851 | 0.87 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
21487 |
0.18 |
| chr5_54734053_54734276 | 0.83 |
SKIV2L2 |
superkiller viralicidic activity 2-like 2 (S. cerevisiae) |
22280 |
0.2 |
| chr11_63530349_63531105 | 0.81 |
ENSG00000264519 |
. |
34816 |
0.12 |
| chr7_95401616_95402462 | 0.80 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
123 |
0.98 |
| chr19_17953136_17953665 | 0.80 |
JAK3 |
Janus kinase 3 |
1856 |
0.22 |
| chr1_113243148_113243677 | 0.79 |
RHOC |
ras homolog family member C |
4131 |
0.12 |
| chr11_106295016_106295231 | 0.79 |
RP11-680E19.1 |
|
160081 |
0.04 |
| chr2_175743189_175743666 | 0.78 |
CHN1 |
chimerin 1 |
30948 |
0.19 |
| chr18_9417287_9417498 | 0.76 |
RALBP1 |
ralA binding protein 1 |
57615 |
0.1 |
| chr12_104194005_104194257 | 0.75 |
RP11-650K20.2 |
|
19257 |
0.13 |
| chr3_11610290_11610777 | 0.75 |
VGLL4 |
vestigial like 4 (Drosophila) |
135 |
0.97 |
| chr5_131560990_131561267 | 0.75 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
1807 |
0.32 |
| chr2_241519242_241519542 | 0.75 |
RNPEPL1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
4473 |
0.13 |
| chr7_101579659_101580126 | 0.75 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr19_41108244_41108427 | 0.75 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
1058 |
0.42 |
| chr2_60757710_60758018 | 0.73 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
22676 |
0.2 |
| chr20_50721484_50721986 | 0.73 |
ZFP64 |
ZFP64 zinc finger protein |
273 |
0.95 |
| chr12_10874089_10875353 | 0.73 |
YBX3 |
Y box binding protein 3 |
1185 |
0.46 |
| chr11_69925187_69925338 | 0.73 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
583 |
0.69 |
| chr15_44486029_44486998 | 0.70 |
FRMD5 |
FERM domain containing 5 |
131 |
0.98 |
| chr9_130860266_130860759 | 0.70 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
71 |
0.93 |
| chr17_80482487_80482690 | 0.68 |
FOXK2 |
forkhead box K2 |
4997 |
0.12 |
| chr12_113860437_113860657 | 0.68 |
SDSL |
serine dehydratase-like |
289 |
0.88 |
| chr5_45163259_45163410 | 0.68 |
MRPS30 |
mitochondrial ribosomal protein S30 |
354307 |
0.01 |
| chr1_203449915_203450277 | 0.67 |
PRELP |
proline/arginine-rich end leucine-rich repeat protein |
5140 |
0.23 |
| chr7_137680969_137681574 | 0.66 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
5522 |
0.22 |
| chr16_88837427_88837668 | 0.65 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
14072 |
0.08 |
| chr3_55520023_55520469 | 0.65 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1085 |
0.48 |
| chr4_25031967_25032301 | 0.65 |
LGI2 |
leucine-rich repeat LGI family, member 2 |
153 |
0.97 |
| chr1_33207592_33208218 | 0.64 |
KIAA1522 |
KIAA1522 |
419 |
0.8 |
| chr1_2144612_2144924 | 0.64 |
AL590822.1 |
Uncharacterized protein |
852 |
0.47 |
| chr1_220101375_220102142 | 0.64 |
SLC30A10 |
solute carrier family 30, member 10 |
186 |
0.95 |
| chr13_99714734_99715210 | 0.63 |
DOCK9 |
dedicator of cytokinesis 9 |
23688 |
0.17 |
| chr1_59043462_59043613 | 0.63 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
371 |
0.88 |
| chr1_205450827_205450978 | 0.62 |
CDK18 |
cyclin-dependent kinase 18 |
22821 |
0.14 |
| chr14_58797643_58798017 | 0.62 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
175 |
0.93 |
| chr5_158521759_158522243 | 0.62 |
EBF1 |
early B-cell factor 1 |
4700 |
0.29 |
| chr13_74784971_74785134 | 0.62 |
KLF12 |
Kruppel-like factor 12 |
76658 |
0.11 |
| chr8_145022099_145022649 | 0.62 |
PLEC |
plectin |
1659 |
0.22 |
| chrX_139846886_139847110 | 0.62 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
19725 |
0.22 |
| chr4_8317573_8317791 | 0.61 |
HTRA3 |
HtrA serine peptidase 3 |
46178 |
0.14 |
| chr20_59829000_59829235 | 0.61 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
1635 |
0.56 |
| chr8_141728080_141728633 | 0.61 |
PTK2 |
protein tyrosine kinase 2 |
98 |
0.98 |
| chr14_55119300_55119828 | 0.60 |
SAMD4A |
sterile alpha motif domain containing 4A |
84927 |
0.09 |
| chr7_42275396_42275690 | 0.60 |
GLI3 |
GLI family zinc finger 3 |
1069 |
0.7 |
| chr16_72882732_72883241 | 0.60 |
ENSG00000251868 |
. |
27095 |
0.17 |
| chr2_228683015_228683311 | 0.60 |
CCL20 |
chemokine (C-C motif) ligand 20 |
4593 |
0.24 |
| chr7_38670068_38671048 | 0.59 |
AMPH |
amphiphysin |
462 |
0.87 |
| chr3_126006504_126006736 | 0.59 |
KLF15 |
Kruppel-like factor 15 |
69665 |
0.08 |
| chr15_65176516_65176787 | 0.59 |
ENSG00000264929 |
. |
15787 |
0.13 |
| chr17_79012089_79012389 | 0.59 |
BAIAP2 |
BAI1-associated protein 2 |
2436 |
0.2 |
| chr4_154064335_154064653 | 0.59 |
TRIM2 |
tripartite motif containing 2 |
9151 |
0.28 |
| chr2_36660395_36660628 | 0.58 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
76897 |
0.1 |
| chr5_98396547_98396924 | 0.58 |
ENSG00000200351 |
. |
124284 |
0.06 |
| chr11_64918203_64918661 | 0.58 |
ENSG00000222477 |
. |
3377 |
0.09 |
| chr11_57089258_57089534 | 0.58 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
275 |
0.83 |
| chr11_95888990_95889540 | 0.57 |
ENSG00000266192 |
. |
185337 |
0.03 |
| chr16_74775111_74775280 | 0.57 |
FA2H |
fatty acid 2-hydroxylase |
28 |
0.98 |
| chr9_124262101_124262557 | 0.57 |
ENSG00000240299 |
. |
5532 |
0.22 |
| chr14_94602985_94603148 | 0.57 |
IFI27L2 |
interferon, alpha-inducible protein 27-like 2 |
6476 |
0.16 |
| chr5_176830840_176831699 | 0.57 |
GRK6 |
G protein-coupled receptor kinase 6 |
138 |
0.91 |
| chr15_70818827_70819034 | 0.57 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
175690 |
0.03 |
| chr19_40871912_40872253 | 0.56 |
PLD3 |
phospholipase D family, member 3 |
181 |
0.9 |
| chr11_126010955_126011106 | 0.56 |
RP11-50B3.4 |
|
67892 |
0.07 |
| chr19_4866504_4867667 | 0.55 |
PLIN3 |
perilipin 3 |
39 |
0.96 |
| chrX_128789131_128789414 | 0.55 |
APLN |
apelin |
339 |
0.92 |
| chr13_24624586_24624810 | 0.54 |
SPATA13 |
spermatogenesis associated 13 |
70754 |
0.1 |
| chr19_34972059_34972243 | 0.54 |
WTIP |
Wilms tumor 1 interacting protein |
277 |
0.89 |
| chr4_74809740_74810515 | 0.54 |
PF4 |
platelet factor 4 |
37714 |
0.11 |
| chr17_20491916_20492779 | 0.54 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr1_159915484_159915973 | 0.54 |
IGSF9 |
immunoglobulin superfamily, member 9 |
342 |
0.79 |
| chr2_185463143_185463634 | 0.54 |
ZNF804A |
zinc finger protein 804A |
295 |
0.95 |
| chr21_28338343_28338505 | 0.53 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
408 |
0.88 |
| chr8_80872484_80872635 | 0.53 |
RP11-26J3.3 |
|
2943 |
0.33 |
| chr7_47610970_47611742 | 0.53 |
TNS3 |
tensin 3 |
9873 |
0.29 |
| chr8_89842231_89842382 | 0.53 |
RP11-586K2.1 |
|
129786 |
0.06 |
| chr22_46402821_46403014 | 0.52 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
29908 |
0.1 |
| chr13_97960259_97960523 | 0.52 |
MBNL2 |
muscleblind-like splicing regulator 2 |
31933 |
0.21 |
| chr3_56817720_56817916 | 0.52 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
8133 |
0.29 |
| chr8_19266488_19266919 | 0.52 |
SH2D4A |
SH2 domain containing 4A |
89602 |
0.1 |
| chr9_130284212_130284363 | 0.52 |
FAM129B |
family with sequence similarity 129, member B |
47080 |
0.09 |
| chr5_66564565_66564999 | 0.51 |
CD180 |
CD180 molecule |
72155 |
0.13 |
| chr22_25372381_25372554 | 0.51 |
KIAA1671 |
KIAA1671 |
23770 |
0.18 |
| chr10_33620140_33620362 | 0.51 |
NRP1 |
neuropilin 1 |
3059 |
0.34 |
| chr17_25707386_25707582 | 0.51 |
TBC1D3P5 |
TBC1 domain family, member 3 pseudogene 5 |
39080 |
0.16 |
| chr11_1897889_1898478 | 0.50 |
LSP1 |
lymphocyte-specific protein 1 |
476 |
0.66 |
| chr4_170235942_170236479 | 0.50 |
SH3RF1 |
SH3 domain containing ring finger 1 |
43954 |
0.19 |
| chr20_50312898_50313325 | 0.50 |
ATP9A |
ATPase, class II, type 9A |
71756 |
0.11 |
| chr17_12887644_12888019 | 0.50 |
ARHGAP44 |
Rho GTPase activating protein 44 |
4406 |
0.18 |
| chr11_86262739_86262907 | 0.50 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
8039 |
0.29 |
| chr13_50194281_50194827 | 0.49 |
ARL11 |
ADP-ribosylation factor-like 11 |
7881 |
0.21 |
| chr8_42063953_42065062 | 0.49 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr21_33784461_33785454 | 0.49 |
EVA1C |
eva-1 homolog C (C. elegans) |
36 |
0.97 |
| chr3_146613212_146613363 | 0.49 |
ENSG00000251800 |
. |
58578 |
0.15 |
| chr14_69419495_69419882 | 0.49 |
ACTN1 |
actinin, alpha 1 |
5423 |
0.25 |
| chr7_115993814_115993965 | 0.48 |
ENSG00000216076 |
. |
7891 |
0.21 |
| chr17_15689649_15690074 | 0.48 |
ENSG00000251829 |
. |
4206 |
0.19 |
| chr17_37036468_37036913 | 0.48 |
LASP1 |
LIM and SH3 protein 1 |
6547 |
0.1 |
| chr2_218843405_218844030 | 0.47 |
TNS1 |
tensin 1 |
60 |
0.79 |
| chr4_159969552_159969736 | 0.47 |
C4orf45 |
chromosome 4 open reading frame 45 |
13311 |
0.17 |
| chr16_4000635_4001034 | 0.47 |
RP11-462G12.1 |
|
389 |
0.84 |
| chr11_66363136_66363287 | 0.47 |
CCS |
copper chaperone for superoxide dismutase |
2499 |
0.13 |
| chr16_74778068_74778306 | 0.47 |
ENSG00000263661 |
. |
2922 |
0.23 |
| chr12_13346530_13346681 | 0.46 |
EMP1 |
epithelial membrane protein 1 |
3045 |
0.33 |
| chr17_38228617_38228812 | 0.46 |
THRA |
thyroid hormone receptor, alpha |
370 |
0.79 |
| chr8_69242814_69243988 | 0.46 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr10_126337337_126337630 | 0.46 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
54711 |
0.13 |
| chr12_53624282_53624433 | 0.45 |
RARG |
retinoic acid receptor, gamma |
1601 |
0.24 |
| chr9_130639317_130639975 | 0.45 |
AK1 |
adenylate kinase 1 |
376 |
0.71 |
| chr2_230135388_230135942 | 0.45 |
PID1 |
phosphotyrosine interaction domain containing 1 |
316 |
0.93 |
| chr7_92324656_92325109 | 0.45 |
ENSG00000206763 |
. |
6246 |
0.27 |
| chr9_4933355_4933539 | 0.45 |
ENSG00000238362 |
. |
34650 |
0.14 |
| chr4_40665217_40665497 | 0.45 |
RBM47 |
RNA binding motif protein 47 |
32465 |
0.19 |
| chr22_46482810_46483184 | 0.45 |
FLJ27365 |
hsa-mir-4763 |
1114 |
0.31 |
| chr16_2085937_2086157 | 0.45 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
2775 |
0.08 |
| chr13_110521954_110522217 | 0.44 |
ENSG00000201161 |
. |
45358 |
0.18 |
| chr17_45982661_45983102 | 0.44 |
SP2 |
Sp2 transcription factor |
9365 |
0.1 |
| chr1_223919032_223919392 | 0.44 |
CAPN2 |
calpain 2, (m/II) large subunit |
19178 |
0.2 |
| chr15_100583304_100583570 | 0.44 |
ENSG00000252957 |
. |
30958 |
0.16 |
| chr1_156659849_156660107 | 0.44 |
NES |
nestin |
12789 |
0.08 |
| chr9_35908111_35908318 | 0.44 |
HRCT1 |
histidine rich carboxyl terminus 1 |
2025 |
0.23 |
| chr4_26374521_26374672 | 0.44 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
10444 |
0.3 |
| chr12_13351879_13352395 | 0.43 |
EMP1 |
epithelial membrane protein 1 |
2417 |
0.37 |
| chr10_11203169_11203338 | 0.43 |
CELF2 |
CUGBP, Elav-like family member 2 |
3740 |
0.26 |
| chr5_176919974_176920125 | 0.43 |
RP11-1334A24.6 |
|
1947 |
0.17 |
| chr2_33361191_33361342 | 0.43 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
1542 |
0.54 |
| chr2_110872063_110872214 | 0.42 |
MALL |
mal, T-cell differentiation protein-like |
1461 |
0.36 |
| chr9_132514262_132514453 | 0.42 |
PTGES |
prostaglandin E synthase |
969 |
0.51 |
| chr3_99356620_99357126 | 0.42 |
COL8A1 |
collagen, type VIII, alpha 1 |
446 |
0.89 |
| chr2_241265147_241265298 | 0.42 |
ENSG00000221412 |
. |
63945 |
0.11 |
| chr3_174158021_174158288 | 0.42 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
623 |
0.85 |
| chr6_158080653_158080804 | 0.42 |
ZDHHC14 |
zinc finger, DHHC-type containing 14 |
66687 |
0.12 |
| chr4_111507244_111507551 | 0.42 |
PITX2 |
paired-like homeodomain 2 |
36810 |
0.19 |
| chr12_6644018_6645022 | 0.42 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
52 |
0.92 |
| chr11_46708891_46709042 | 0.42 |
ARHGAP1 |
Rho GTPase activating protein 1 |
8683 |
0.13 |
| chr9_66486862_66487013 | 0.42 |
ENSG00000202474 |
. |
28675 |
0.27 |
| chr10_79686888_79687096 | 0.42 |
DLG5-AS1 |
DLG5 antisense RNA 1 |
138 |
0.87 |
| chr5_32193714_32193865 | 0.41 |
GOLPH3 |
golgi phosphoprotein 3 (coat-protein) |
19333 |
0.18 |
| chr3_10057070_10057667 | 0.41 |
EMC3 |
ER membrane protein complex subunit 3 |
4568 |
0.11 |
| chr11_59523287_59523695 | 0.41 |
STX3 |
syntaxin 3 |
566 |
0.57 |
| chr12_32050718_32050938 | 0.41 |
ENSG00000252584 |
. |
17813 |
0.2 |
| chr16_10688359_10688510 | 0.41 |
EMP2 |
epithelial membrane protein 2 |
13879 |
0.17 |
| chr10_86068772_86068923 | 0.41 |
CCSER2 |
coiled-coil serine-rich protein 2 |
19495 |
0.2 |
| chr15_41055526_41056933 | 0.41 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
11 |
0.96 |
| chr5_106878314_106878602 | 0.40 |
EFNA5 |
ephrin-A5 |
127870 |
0.06 |
| chr1_56943511_56943702 | 0.40 |
ENSG00000223307 |
. |
100516 |
0.08 |
| chr2_159825200_159826543 | 0.40 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
688 |
0.73 |
| chr19_16936008_16936179 | 0.40 |
SIN3B |
SIN3 transcription regulator family member B |
4118 |
0.16 |
| chr1_212420607_212420921 | 0.40 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
38115 |
0.14 |
| chr7_143078673_143079796 | 0.40 |
ZYX |
zyxin |
234 |
0.85 |
| chr17_1014745_1015083 | 0.40 |
ABR |
active BCR-related |
2574 |
0.31 |
| chr12_106531293_106531501 | 0.40 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
2414 |
0.35 |
| chr14_51338929_51339207 | 0.40 |
ABHD12B |
abhydrolase domain containing 12B |
190 |
0.93 |
| chr8_8749576_8750657 | 0.40 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
1039 |
0.53 |
| chr2_161151049_161151328 | 0.39 |
ITGB6 |
integrin, beta 6 |
94373 |
0.08 |
| chr4_175344307_175344635 | 0.39 |
ENSG00000265846 |
. |
475 |
0.88 |
| chr8_120428169_120429285 | 0.39 |
NOV |
nephroblastoma overexpressed |
181 |
0.96 |
| chr2_167230570_167230946 | 0.39 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
1739 |
0.43 |
| chr19_2578858_2579047 | 0.39 |
CTC-265F19.2 |
|
32908 |
0.12 |
| chr1_17519453_17519604 | 0.39 |
PADI1 |
peptidyl arginine deiminase, type I |
12093 |
0.17 |
| chr7_149193458_149193685 | 0.39 |
ZNF746 |
zinc finger protein 746 |
1253 |
0.51 |
| chr7_2439740_2440019 | 0.39 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
3344 |
0.25 |
| chr2_237086905_237087412 | 0.39 |
GBX2 |
gastrulation brain homeobox 2 |
10146 |
0.15 |
| chr7_55087156_55087307 | 0.38 |
EGFR |
epidermal growth factor receptor |
420 |
0.91 |
| chr7_130013324_130013755 | 0.38 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
6641 |
0.14 |
| chr14_102094993_102095193 | 0.38 |
DIO3 |
deiodinase, iodothyronine, type III |
67405 |
0.09 |
| chr1_99729766_99730318 | 0.38 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
194 |
0.97 |
| chr5_146796712_146796863 | 0.38 |
CTB-108O6.2 |
|
15464 |
0.22 |
| chrX_53309975_53310241 | 0.38 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
688 |
0.69 |
| chr7_28527889_28528184 | 0.38 |
CREB5 |
cAMP responsive element binding protein 5 |
2712 |
0.43 |
| chr18_9647997_9648175 | 0.38 |
ENSG00000212572 |
. |
30016 |
0.14 |
| chr3_13060724_13060906 | 0.38 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
32279 |
0.22 |
| chr2_14775112_14775752 | 0.38 |
AC011897.1 |
Uncharacterized protein |
217 |
0.95 |
| chr17_25886913_25887375 | 0.37 |
KSR1 |
kinase suppressor of ras 1 |
22803 |
0.19 |
| chr9_32741891_32742042 | 0.37 |
ENSG00000221696 |
. |
22328 |
0.18 |
| chr9_21964953_21966022 | 0.37 |
C9orf53 |
chromosome 9 open reading frame 53 |
1650 |
0.31 |
| chr8_77316107_77316384 | 0.37 |
ENSG00000222231 |
. |
138278 |
0.05 |
| chr4_41259507_41259760 | 0.37 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
671 |
0.51 |
| chr2_171822769_171822920 | 0.37 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
37013 |
0.16 |
| chr2_11508334_11508566 | 0.37 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
23739 |
0.2 |
| chr22_46330885_46331075 | 0.36 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
37615 |
0.13 |
| chr1_956379_957022 | 0.36 |
AGRN |
agrin |
1197 |
0.27 |
| chr2_61407466_61407663 | 0.36 |
AHSA2 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
2900 |
0.22 |
| chr17_78044749_78045392 | 0.36 |
CCDC40 |
coiled-coil domain containing 40 |
12346 |
0.15 |
| chr3_196133770_196133921 | 0.36 |
ENSG00000243339 |
. |
7063 |
0.11 |
| chr20_30429794_30429945 | 0.36 |
FOXS1 |
forkhead box S1 |
3551 |
0.16 |
| chr3_45052751_45053653 | 0.36 |
CLEC3B |
C-type lectin domain family 3, member B |
14473 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.2 | 1.1 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.1 | 0.4 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 | 0.4 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.4 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.8 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.3 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 0.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.1 | 0.2 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.2 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.7 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.1 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.7 | GO:0060438 | trachea development(GO:0060438) |
| 0.1 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 0.3 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.4 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.1 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.2 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.4 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.3 | GO:0007004 | telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.3 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.0 | 0.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0009309 | amine biosynthetic process(GO:0009309) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.1 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.0 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 3.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.2 | 1.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 0.6 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.2 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.2 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |