Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
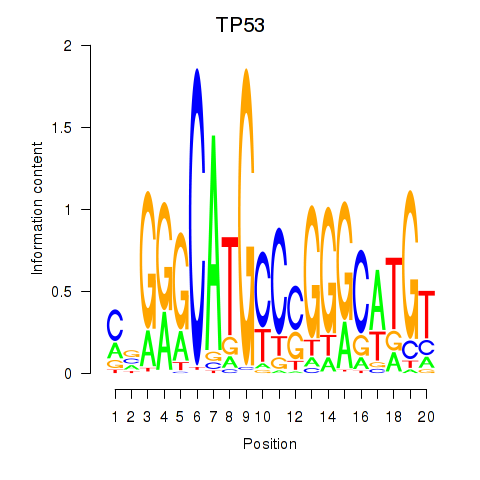
Results for TP53
Z-value: 1.49
Transcription factors associated with TP53
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP53
|
ENSG00000141510.11 | tumor protein p53 |
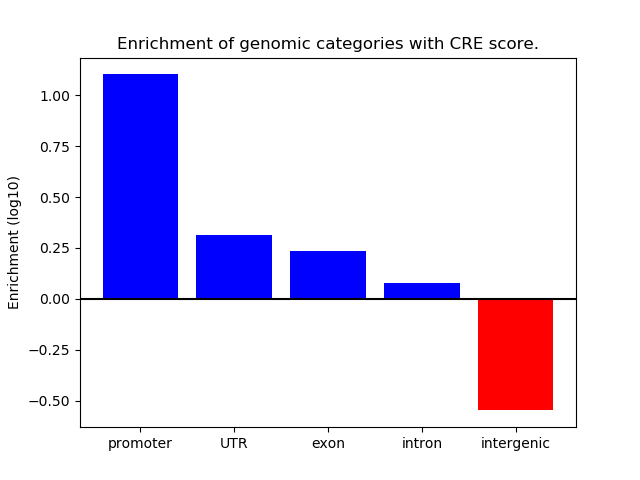
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_7590649_7590937 | TP53 | 6 | 0.494120 | -0.29 | 4.6e-01 | Click! |
| chr17_7590956_7591177 | TP53 | 210 | 0.564802 | -0.21 | 5.8e-01 | Click! |
| chr17_7590042_7590608 | TP53 | 420 | 0.348590 | -0.12 | 7.6e-01 | Click! |
Activity of the TP53 motif across conditions
Conditions sorted by the z-value of the TP53 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
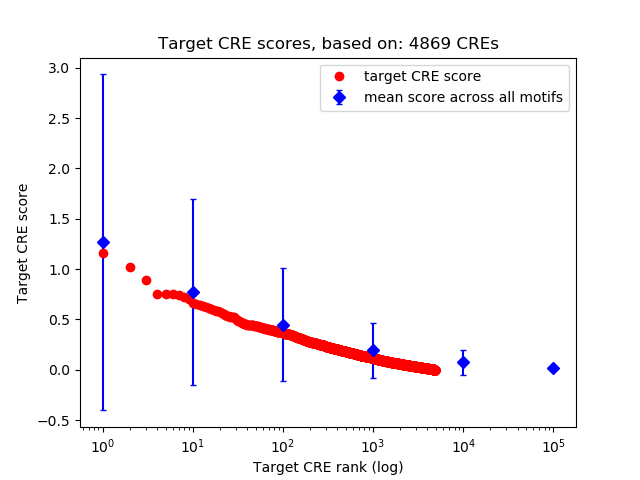
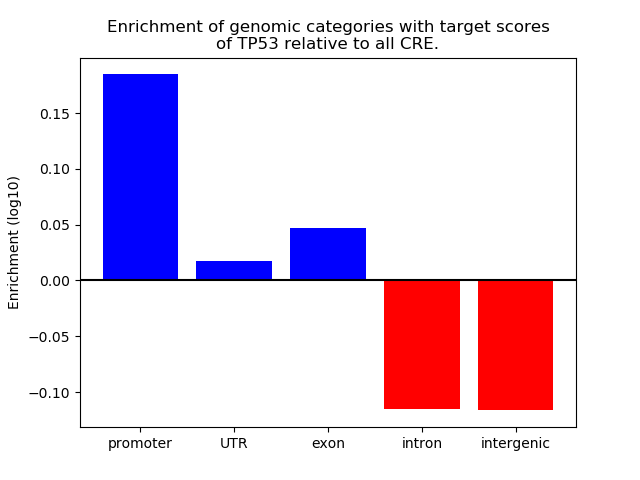
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_72439858_72440009 | 1.16 |
TRIM74 |
tripartite motif containing 74 |
64 |
0.95 |
| chr7_75024883_75025034 | 1.02 |
TRIM73 |
tripartite motif containing 73 |
55 |
0.84 |
| chr21_36077269_36077677 | 0.89 |
CLIC6 |
chloride intracellular channel 6 |
35785 |
0.17 |
| chr1_116381723_116382451 | 0.75 |
NHLH2 |
nescient helix loop helix 2 |
1275 |
0.57 |
| chr1_16950904_16951055 | 0.75 |
NBPF1 |
neuroblastoma breakpoint family, member 1 |
10997 |
0.14 |
| chr10_5566796_5566947 | 0.75 |
CALML3 |
calmodulin-like 3 |
53 |
0.92 |
| chr9_110011481_110011632 | 0.75 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
33862 |
0.22 |
| chr4_1760272_1760423 | 0.72 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
30216 |
0.12 |
| chrX_113309122_113309273 | 0.70 |
ENSG00000212611 |
. |
48936 |
0.16 |
| chr7_2019823_2019974 | 0.66 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
528 |
0.86 |
| chr11_706564_706815 | 0.65 |
EPS8L2 |
EPS8-like 2 |
60 |
0.94 |
| chr19_45313552_45313703 | 0.65 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
1249 |
0.33 |
| chr9_131865142_131865293 | 0.64 |
ENSG00000221521 |
. |
4213 |
0.12 |
| chr9_129263535_129263686 | 0.63 |
ENSG00000221768 |
. |
29660 |
0.17 |
| chr17_35718321_35718472 | 0.62 |
ACACA |
acetyl-CoA carboxylase alpha |
2337 |
0.25 |
| chr11_1328574_1328800 | 0.60 |
TOLLIP |
toll interacting protein |
2162 |
0.23 |
| chr4_54968164_54968315 | 0.59 |
ENSG00000221219 |
. |
1584 |
0.3 |
| chr9_132094734_132094885 | 0.59 |
ENSG00000242281 |
. |
37931 |
0.16 |
| chr5_1004071_1004222 | 0.58 |
ENSG00000221244 |
. |
156 |
0.93 |
| chr16_29262868_29263019 | 0.58 |
RP11-231C14.6 |
|
60728 |
0.09 |
| chr10_119303143_119303333 | 0.57 |
EMX2 |
empty spiracles homeobox 2 |
729 |
0.61 |
| chr22_20887250_20887401 | 0.55 |
MED15 |
mediator complex subunit 15 |
8763 |
0.12 |
| chr18_28622614_28622900 | 0.54 |
DSC3 |
desmocollin 3 |
24 |
0.98 |
| chr22_46480425_46480827 | 0.54 |
FLJ27365 |
hsa-mir-4763 |
1257 |
0.28 |
| chr4_157692546_157692809 | 0.54 |
RP11-154F14.2 |
|
69834 |
0.11 |
| chr17_4463313_4463464 | 0.53 |
GGT6 |
gamma-glutamyltransferase 6 |
158 |
0.92 |
| chr15_41913046_41914152 | 0.53 |
MGA |
MGA, MAX dimerization protein |
135 |
0.96 |
| chr16_902323_902474 | 0.52 |
LA16c-306A4.1 |
|
31382 |
0.08 |
| chr16_67213263_67213414 | 0.52 |
KIAA0895L |
KIAA0895-like |
627 |
0.43 |
| chr1_177139870_177140730 | 0.51 |
BRINP2 |
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
333 |
0.89 |
| chr1_145543160_145543350 | 0.50 |
RP11-315I20.3 |
|
1026 |
0.34 |
| chr16_57654156_57654307 | 0.49 |
GPR56 |
G protein-coupled receptor 56 |
215 |
0.92 |
| chr11_119567368_119567519 | 0.48 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
31820 |
0.14 |
| chr11_8111770_8111921 | 0.47 |
TUB |
tubby bipartite transcription factor |
8936 |
0.16 |
| chr3_124913489_124913640 | 0.47 |
SLC12A8 |
solute carrier family 12, member 8 |
2025 |
0.37 |
| chr2_214334_214485 | 0.46 |
AC079779.7 |
|
16840 |
0.18 |
| chr20_61003066_61003217 | 0.46 |
RBBP8NL |
RBBP8 N-terminal like |
552 |
0.7 |
| chr19_19446684_19446835 | 0.46 |
MAU2 |
MAU2 sister chromatid cohesion factor |
1256 |
0.36 |
| chr3_55520023_55520469 | 0.45 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1085 |
0.48 |
| chr22_46474299_46474631 | 0.45 |
FLJ27365 |
hsa-mir-4763 |
1727 |
0.2 |
| chr16_54459980_54460131 | 0.45 |
ENSG00000264079 |
. |
123253 |
0.06 |
| chr11_129061773_129061924 | 0.44 |
ARHGAP32 |
Rho GTPase activating protein 32 |
245 |
0.94 |
| chr6_46620180_46620625 | 0.44 |
CYP39A1 |
cytochrome P450, family 39, subfamily A, polypeptide 1 |
121 |
0.71 |
| chr12_54519921_54520255 | 0.44 |
ENSG00000202146 |
. |
27002 |
0.09 |
| chr2_232544960_232545111 | 0.44 |
MGC4771 |
|
26586 |
0.13 |
| chr7_67474743_67474894 | 0.44 |
ENSG00000265600 |
. |
653151 |
0.0 |
| chr8_21959623_21959774 | 0.44 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
7212 |
0.11 |
| chr14_104638147_104638298 | 0.44 |
KIF26A |
kinesin family member 26A |
32845 |
0.13 |
| chr6_34122459_34122617 | 0.44 |
GRM4 |
glutamate receptor, metabotropic 4 |
861 |
0.68 |
| chr14_76843049_76843200 | 0.44 |
ENSG00000263880 |
. |
21772 |
0.18 |
| chr19_28285219_28285519 | 0.43 |
AC005758.1 |
|
124482 |
0.06 |
| chr16_57687976_57688127 | 0.43 |
GPR56 |
G protein-coupled receptor 56 |
908 |
0.48 |
| chr22_35715502_35715653 | 0.43 |
TOM1 |
target of myb1 (chicken) |
8522 |
0.16 |
| chr2_137522313_137522464 | 0.42 |
THSD7B |
thrombospondin, type I, domain containing 7B |
727 |
0.8 |
| chr8_144960354_144960505 | 0.42 |
EPPK1 |
epiplakin 1 |
7797 |
0.11 |
| chr18_45599789_45599940 | 0.42 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
32370 |
0.22 |
| chr17_37031531_37031718 | 0.42 |
LASP1 |
LIM and SH3 protein 1 |
1481 |
0.23 |
| chr17_37760675_37760938 | 0.42 |
NEUROD2 |
neuronal differentiation 2 |
3390 |
0.17 |
| chr8_10588268_10588485 | 0.41 |
SOX7 |
SRY (sex determining region Y)-box 7 |
354 |
0.78 |
| chr12_54391177_54391923 | 0.41 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
981 |
0.22 |
| chrX_153029718_153029902 | 0.41 |
PLXNB3 |
plexin B3 |
84 |
0.93 |
| chr11_75152424_75152575 | 0.41 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
268 |
0.88 |
| chr14_105332332_105332761 | 0.41 |
CEP170B |
centrosomal protein 170B |
892 |
0.57 |
| chr1_38465984_38466787 | 0.41 |
FHL3 |
four and a half LIM domains 3 |
4792 |
0.14 |
| chr4_96461530_96461681 | 0.41 |
UNC5C |
unc-5 homolog C (C. elegans) |
8518 |
0.25 |
| chr11_65666484_65666730 | 0.41 |
FOSL1 |
FOS-like antigen 1 |
1283 |
0.23 |
| chr11_64073389_64074392 | 0.40 |
ESRRA |
estrogen-related receptor alpha |
28 |
0.92 |
| chr8_96237876_96238027 | 0.40 |
C8orf37 |
chromosome 8 open reading frame 37 |
43478 |
0.17 |
| chr19_18475720_18476136 | 0.40 |
PGPEP1 |
pyroglutamyl-peptidase I |
7612 |
0.09 |
| chr8_69242814_69243988 | 0.40 |
C8orf34 |
chromosome 8 open reading frame 34 |
56 |
0.82 |
| chr19_35589604_35589755 | 0.40 |
AC020907.1 |
Uncharacterized protein |
7194 |
0.1 |
| chr1_113392388_113392874 | 0.40 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
85979 |
0.07 |
| chr1_6085866_6086187 | 0.40 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
334 |
0.88 |
| chr7_129420780_129420931 | 0.40 |
ENSG00000207691 |
. |
6001 |
0.16 |
| chr3_182879726_182880740 | 0.40 |
LAMP3 |
lysosomal-associated membrane protein 3 |
430 |
0.86 |
| chr19_56652318_56652782 | 0.39 |
ZNF444 |
zinc finger protein 444 |
6 |
0.96 |
| chr17_79056077_79056228 | 0.39 |
BAIAP2 |
BAI1-associated protein 2 |
4109 |
0.15 |
| chr21_44872011_44872162 | 0.39 |
SIK1 |
salt-inducible kinase 1 |
25078 |
0.24 |
| chr1_107682797_107683388 | 0.39 |
NTNG1 |
netrin G1 |
350 |
0.93 |
| chr12_26379464_26379833 | 0.39 |
SSPN |
sarcospan |
31042 |
0.17 |
| chr16_89118855_89119113 | 0.39 |
ACSF3 |
acyl-CoA synthetase family member 3 |
35799 |
0.13 |
| chr22_46861776_46861927 | 0.39 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
69340 |
0.1 |
| chr1_221166154_221166305 | 0.38 |
HLX |
H2.0-like homeobox |
111645 |
0.06 |
| chr19_30159686_30159837 | 0.38 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
1980 |
0.38 |
| chr10_99309798_99309949 | 0.38 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
22325 |
0.1 |
| chr22_43523383_43523534 | 0.38 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
15512 |
0.13 |
| chr9_139493449_139493600 | 0.38 |
ENSG00000252440 |
. |
3443 |
0.14 |
| chr4_48988009_48988286 | 0.38 |
CWH43 |
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) |
117 |
0.97 |
| chr19_4010009_4010213 | 0.37 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
2467 |
0.16 |
| chr19_17905710_17906188 | 0.37 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
4 |
0.96 |
| chr1_201253218_201253369 | 0.37 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
462 |
0.83 |
| chr12_56546304_56546955 | 0.37 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
238 |
0.78 |
| chr3_187491387_187491633 | 0.37 |
BCL6 |
B-cell CLL/lymphoma 6 |
27995 |
0.2 |
| chr20_22807182_22807333 | 0.37 |
ENSG00000265151 |
. |
93318 |
0.08 |
| chr20_48766370_48766669 | 0.37 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
3655 |
0.2 |
| chr11_2824838_2824989 | 0.37 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
57885 |
0.09 |
| chr7_738673_738824 | 0.37 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
7317 |
0.21 |
| chr18_8706613_8707165 | 0.37 |
SOGA2 |
SOGA family member 2 |
520 |
0.55 |
| chr1_149858479_149859475 | 0.37 |
HIST2H2AC |
histone cluster 2, H2ac |
452 |
0.26 |
| chr2_213793107_213793258 | 0.37 |
ENSG00000266354 |
. |
2201 |
0.44 |
| chr11_2230168_2230319 | 0.36 |
ENSG00000265258 |
. |
35950 |
0.09 |
| chr22_46465997_46466532 | 0.36 |
RP6-109B7.4 |
|
493 |
0.65 |
| chr19_41847932_41848083 | 0.36 |
TGFB1 |
transforming growth factor, beta 1 |
72 |
0.94 |
| chr10_119301594_119301816 | 0.36 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
125 |
0.68 |
| chr1_154785958_154786192 | 0.36 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
46244 |
0.11 |
| chr16_787839_787990 | 0.36 |
NARFL |
nuclear prelamin A recognition factor-like |
25 |
0.92 |
| chr3_46671629_46671864 | 0.36 |
ENSG00000251967 |
. |
20254 |
0.13 |
| chr22_50630477_50630948 | 0.36 |
RP3-402G11.25 |
|
119 |
0.86 |
| chr6_158181766_158181950 | 0.36 |
SNX9 |
sorting nexin 9 |
62438 |
0.11 |
| chr13_102068774_102068925 | 0.36 |
NALCN |
sodium leak channel, non-selective |
65 |
0.98 |
| chr21_36216765_36216916 | 0.36 |
RUNX1 |
runt-related transcription factor 1 |
42640 |
0.19 |
| chr2_75787847_75788044 | 0.36 |
EVA1A |
eva-1 homolog A (C. elegans) |
94 |
0.97 |
| chr11_6340744_6340939 | 0.36 |
PRKCDBP |
protein kinase C, delta binding protein |
921 |
0.53 |
| chr5_131729150_131729301 | 0.36 |
C5orf56 |
chromosome 5 open reading frame 56 |
17103 |
0.11 |
| chr1_16480833_16481011 | 0.36 |
RP11-276H7.2 |
|
784 |
0.46 |
| chr8_22576706_22576857 | 0.35 |
EGR3 |
early growth response 3 |
25966 |
0.12 |
| chr11_12132812_12133164 | 0.35 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
850 |
0.73 |
| chr1_45806125_45806469 | 0.35 |
MUTYH |
mutY homolog |
155 |
0.84 |
| chr6_44030171_44030322 | 0.35 |
RP5-1120P11.1 |
|
12143 |
0.17 |
| chr5_149315304_149315455 | 0.35 |
ENSG00000200334 |
. |
4315 |
0.18 |
| chr9_98225406_98225667 | 0.35 |
PTCH1 |
patched 1 |
17231 |
0.18 |
| chr9_130831197_130832052 | 0.35 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
1052 |
0.31 |
| chr10_105842975_105843126 | 0.35 |
COL17A1 |
collagen, type XVII, alpha 1 |
2486 |
0.27 |
| chr1_6418349_6418500 | 0.35 |
ACOT7 |
acyl-CoA thioesterase 7 |
980 |
0.49 |
| chr16_70624804_70624955 | 0.34 |
IL34 |
interleukin 34 |
11067 |
0.15 |
| chr19_35609321_35609472 | 0.34 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
38 |
0.95 |
| chr9_130186748_130187435 | 0.34 |
ZNF79 |
zinc finger protein 79 |
374 |
0.8 |
| chr17_73567230_73567381 | 0.34 |
MYO15B |
myosin XVB pseudogene |
19553 |
0.1 |
| chr11_34175761_34176017 | 0.34 |
ENSG00000201867 |
. |
34832 |
0.18 |
| chr10_134211385_134212056 | 0.34 |
PWWP2B |
PWWP domain containing 2B |
1018 |
0.54 |
| chr20_61002159_61002310 | 0.34 |
RBBP8NL |
RBBP8 N-terminal like |
355 |
0.83 |
| chr10_106034668_106034899 | 0.34 |
GSTO2 |
glutathione S-transferase omega 2 |
134 |
0.94 |
| chr7_99754005_99754156 | 0.34 |
C7orf43 |
chromosome 7 open reading frame 43 |
17 |
0.71 |
| chr21_46824152_46824303 | 0.34 |
COL18A1 |
collagen, type XVIII, alpha 1 |
825 |
0.56 |
| chr1_2345264_2345415 | 0.34 |
PEX10 |
peroxisomal biogenesis factor 10 |
103 |
0.94 |
| chr3_48468926_48469077 | 0.33 |
PLXNB1 |
plexin B1 |
1871 |
0.18 |
| chr19_16396913_16397119 | 0.33 |
CTD-2562J15.6 |
|
7370 |
0.17 |
| chr3_126006504_126006736 | 0.33 |
KLF15 |
Kruppel-like factor 15 |
69665 |
0.08 |
| chr6_161454686_161454957 | 0.33 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
11595 |
0.25 |
| chr10_16483414_16483776 | 0.33 |
PTER |
phosphotriesterase related |
4612 |
0.25 |
| chr1_9241794_9242495 | 0.33 |
ENSG00000207865 |
. |
30308 |
0.15 |
| chr17_32947230_32947381 | 0.33 |
TMEM132E |
transmembrane protein 132E |
39537 |
0.17 |
| chr13_77343113_77343264 | 0.33 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
117337 |
0.06 |
| chr19_4374282_4374693 | 0.32 |
SH3GL1 |
SH3-domain GRB2-like 1 |
5802 |
0.08 |
| chr5_178487316_178488173 | 0.32 |
ZNF354C |
zinc finger protein 354C |
328 |
0.9 |
| chr16_69134943_69135094 | 0.32 |
HAS3 |
hyaluronan synthase 3 |
4449 |
0.18 |
| chr1_234793534_234793917 | 0.32 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
48454 |
0.13 |
| chr15_40532500_40532651 | 0.32 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
148 |
0.93 |
| chr8_41166402_41166711 | 0.32 |
SFRP1 |
secreted frizzled-related protein 1 |
460 |
0.8 |
| chr6_33547162_33547696 | 0.32 |
BAK1 |
BCL2-antagonist/killer 1 |
552 |
0.67 |
| chr6_53493330_53493549 | 0.32 |
RP11-345L23.1 |
|
261 |
0.92 |
| chr20_22565035_22565186 | 0.32 |
FOXA2 |
forkhead box A2 |
9 |
0.99 |
| chr7_4046197_4046504 | 0.31 |
SDK1 |
sidekick cell adhesion molecule 1 |
55798 |
0.18 |
| chr18_67959305_67959456 | 0.31 |
SOCS6 |
suppressor of cytokine signaling 6 |
3243 |
0.3 |
| chr2_220154476_220154679 | 0.31 |
ENSG00000207647 |
. |
4345 |
0.09 |
| chr8_102039581_102039732 | 0.31 |
ENSG00000252736 |
. |
64385 |
0.1 |
| chr6_36089175_36089326 | 0.31 |
MAPK13 |
mitogen-activated protein kinase 13 |
8848 |
0.18 |
| chr8_144682749_144682900 | 0.31 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
1113 |
0.23 |
| chr1_55525795_55525946 | 0.31 |
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
19975 |
0.19 |
| chr1_61668583_61669016 | 0.31 |
RP4-802A10.1 |
|
78394 |
0.11 |
| chr11_102138737_102139426 | 0.31 |
RP11-864G5.3 |
|
37428 |
0.13 |
| chr21_32715523_32716836 | 0.31 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
415 |
0.91 |
| chr1_1005420_1005571 | 0.31 |
RNF223 |
ring finger protein 223 |
4192 |
0.11 |
| chr22_32079811_32079962 | 0.30 |
PISD |
phosphatidylserine decarboxylase |
21468 |
0.16 |
| chr15_76442977_76443390 | 0.30 |
RP11-593F23.1 |
|
30644 |
0.19 |
| chr6_24402907_24403844 | 0.30 |
MRS2 |
MRS2 magnesium transporter |
45 |
0.98 |
| chr4_1324444_1324595 | 0.30 |
UVSSA |
UV-stimulated scaffold protein A |
16535 |
0.12 |
| chr6_166581874_166582172 | 0.30 |
T |
T, brachyury homolog (mouse) |
84 |
0.98 |
| chr6_42248792_42248943 | 0.30 |
MRPS10 |
mitochondrial ribosomal protein S10 |
63264 |
0.1 |
| chr5_9467649_9467848 | 0.30 |
CTD-2201E9.2 |
|
55542 |
0.13 |
| chr11_1783983_1784134 | 0.30 |
CTSD |
cathepsin D |
425 |
0.53 |
| chr1_110776481_110776632 | 0.30 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
22434 |
0.12 |
| chr3_44040787_44041403 | 0.29 |
ENSG00000252980 |
. |
71484 |
0.12 |
| chr4_62066317_62066605 | 0.29 |
LPHN3 |
latrophilin 3 |
515 |
0.89 |
| chr3_197121264_197121415 | 0.29 |
ENSG00000238491 |
. |
54917 |
0.13 |
| chr14_105443453_105443604 | 0.29 |
AHNAK2 |
AHNAK nucleoprotein 2 |
1166 |
0.43 |
| chr19_8559121_8559517 | 0.29 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
8176 |
0.12 |
| chr6_150285486_150285915 | 0.29 |
ULBP1 |
UL16 binding protein 1 |
557 |
0.69 |
| chr3_6343229_6343380 | 0.29 |
GRM7 |
glutamate receptor, metabotropic 7 |
468452 |
0.01 |
| chr10_99443919_99444098 | 0.29 |
AVPI1 |
arginine vasopressin-induced 1 |
3072 |
0.2 |
| chr3_50377359_50378278 | 0.29 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
454 |
0.47 |
| chr1_205419612_205419763 | 0.29 |
LEMD1 |
LEM domain containing 1 |
628 |
0.66 |
| chr19_626975_627126 | 0.29 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
3118 |
0.11 |
| chr8_10587716_10588163 | 0.29 |
SOX7 |
SRY (sex determining region Y)-box 7 |
83 |
0.93 |
| chr6_167171492_167171681 | 0.29 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
187 |
0.97 |
| chr20_23617810_23618640 | 0.28 |
CST3 |
cystatin C |
357 |
0.87 |
| chr17_29887299_29887674 | 0.28 |
ENSG00000207614 |
. |
471 |
0.69 |
| chr14_99713029_99713259 | 0.28 |
AL109767.1 |
|
16141 |
0.21 |
| chr8_145020311_145020609 | 0.28 |
PLEC |
plectin |
113 |
0.91 |
| chr16_85751517_85751668 | 0.28 |
C16orf74 |
chromosome 16 open reading frame 74 |
7136 |
0.13 |
| chr10_125751436_125751814 | 0.28 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
54616 |
0.13 |
| chr19_11533070_11533503 | 0.28 |
RGL3 |
ral guanine nucleotide dissociation stimulator-like 3 |
3268 |
0.1 |
| chr15_99995321_99995472 | 0.28 |
MEF2A |
myocyte enhancer factor 2A |
21974 |
0.18 |
| chr11_129856033_129856184 | 0.28 |
PRDM10 |
PR domain containing 10 |
16572 |
0.23 |
| chr11_125133143_125133294 | 0.28 |
RP11-687M24.8 |
|
3476 |
0.23 |
| chr8_144951231_144951382 | 0.28 |
EPPK1 |
epiplakin 1 |
1326 |
0.28 |
| chr12_6485624_6485886 | 0.28 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
770 |
0.42 |
| chr8_21763339_21763611 | 0.28 |
DOK2 |
docking protein 2, 56kDa |
7699 |
0.2 |
| chr4_25915833_25916134 | 0.28 |
SMIM20 |
small integral membrane protein 20 |
52 |
0.98 |
| chr16_15360582_15360733 | 0.28 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
103269 |
0.05 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.6 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.4 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.1 | 0.5 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 0.2 | GO:0060068 | vagina development(GO:0060068) |
| 0.1 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.4 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.3 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.1 | 0.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.3 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.2 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.4 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 0.2 | GO:0090192 | regulation of glomerulus development(GO:0090192) |
| 0.1 | 0.1 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.1 | 0.2 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.1 | 0.2 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 | 0.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.2 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0090594 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.2 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.2 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.3 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.3 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.4 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0061162 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0060487 | lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0043247 | telomere capping(GO:0016233) protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.5 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0052308 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.2 | GO:0038061 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0042508 | tyrosine phosphorylation of Stat1 protein(GO:0042508) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0060761 | negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.0 | 0.0 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translation in response to stress(GO:0032055) negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0072203 | cell proliferation involved in metanephros development(GO:0072203) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.1 | GO:0001662 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0032459 | regulation of protein oligomerization(GO:0032459) |
| 0.0 | 0.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0007440 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.2 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) otic vesicle development(GO:0071599) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.1 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0032155 | cell division site(GO:0032153) cell division site part(GO:0032155) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 0.1 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.1 | 0.1 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME SEMAPHORIN INTERACTIONS | Genes involved in Semaphorin interactions |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |