Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
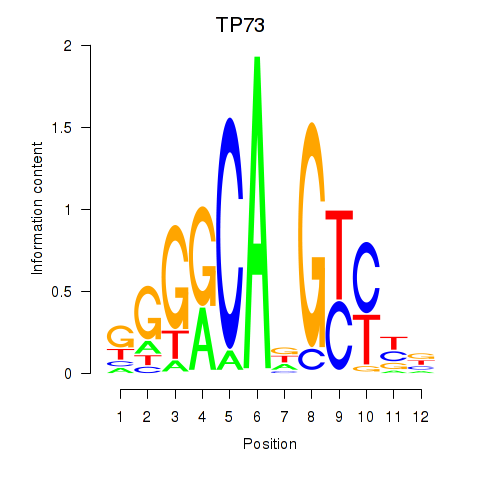
Results for TP73
Z-value: 0.73
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_3619377_3619528 | TP73 | 4830 | 0.130937 | 0.78 | 1.3e-02 | Click! |
| chr1_3592311_3592462 | TP73 | 6544 | 0.121792 | 0.75 | 1.9e-02 | Click! |
| chr1_3568591_3568742 | TP73 | 418 | 0.570948 | 0.57 | 1.1e-01 | Click! |
| chr1_3619628_3619779 | TP73 | 5081 | 0.128982 | 0.53 | 1.4e-01 | Click! |
| chr1_3620270_3620421 | TP73 | 5723 | 0.124950 | 0.51 | 1.6e-01 | Click! |
Activity of the TP73 motif across conditions
Conditions sorted by the z-value of the TP73 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
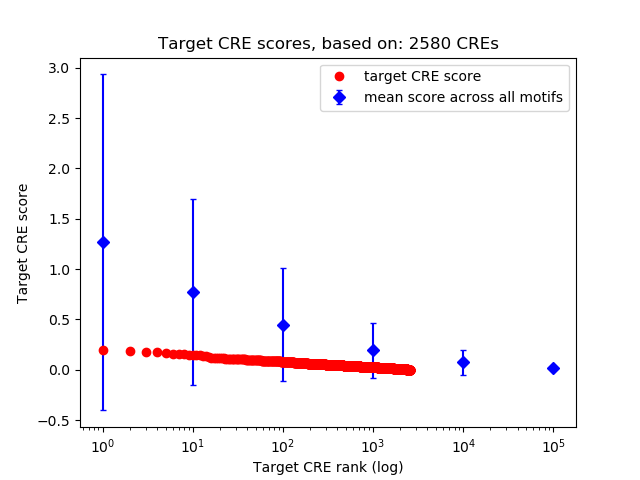
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_129846734_129847253 | 0.20 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1159 |
0.62 |
| chr1_27669122_27669371 | 0.19 |
SYTL1 |
synaptotagmin-like 1 |
473 |
0.71 |
| chr11_64107928_64108112 | 0.18 |
CCDC88B |
coiled-coil domain containing 88B |
325 |
0.74 |
| chr17_1000098_1000249 | 0.17 |
ABR |
active BCR-related |
12167 |
0.2 |
| chr1_26872928_26873171 | 0.17 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
706 |
0.6 |
| chr17_8869162_8869418 | 0.16 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
266 |
0.94 |
| chr22_39493202_39493353 | 0.16 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
3 |
0.97 |
| chr7_45017320_45017514 | 0.15 |
MYO1G |
myosin IG |
1280 |
0.35 |
| chr19_1038413_1038573 | 0.15 |
AC011558.5 |
|
570 |
0.49 |
| chr10_135089035_135089208 | 0.15 |
ADAM8 |
ADAM metallopeptidase domain 8 |
1233 |
0.29 |
| chr20_47439188_47439680 | 0.15 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
4986 |
0.3 |
| chr7_67474743_67474894 | 0.14 |
ENSG00000265600 |
. |
653151 |
0.0 |
| chr20_62708665_62709256 | 0.14 |
RGS19 |
regulator of G-protein signaling 19 |
1885 |
0.17 |
| chr3_195806787_195806938 | 0.14 |
TFRC |
transferrin receptor |
2099 |
0.29 |
| chr1_26872331_26872899 | 0.12 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
272 |
0.88 |
| chr17_43301591_43301923 | 0.12 |
CTD-2020K17.1 |
|
2168 |
0.16 |
| chr19_13207805_13208124 | 0.12 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
5717 |
0.11 |
| chr17_3818740_3818909 | 0.12 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
970 |
0.51 |
| chr22_46684122_46684292 | 0.12 |
GTSE1 |
G-2 and S-phase expressed 1 |
8431 |
0.15 |
| chr7_50351468_50351619 | 0.11 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
3225 |
0.36 |
| chr12_122250872_122251023 | 0.11 |
SETD1B |
SET domain containing 1B |
8309 |
0.15 |
| chr20_826966_827117 | 0.11 |
FAM110A |
family with sequence similarity 110, member A |
1274 |
0.53 |
| chr10_73847489_73847640 | 0.11 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
522 |
0.83 |
| chr10_135073019_135073240 | 0.11 |
MIR202HG |
MIR202 host gene (non-protein coding) |
11734 |
0.1 |
| chr10_49894087_49894238 | 0.11 |
WDFY4 |
WDFY family member 4 |
1241 |
0.52 |
| chr19_5804658_5804817 | 0.11 |
DUS3L |
dihydrouridine synthase 3-like (S. cerevisiae) |
13488 |
0.08 |
| chr12_9912735_9912912 | 0.11 |
CD69 |
CD69 molecule |
674 |
0.66 |
| chr7_1551351_1551591 | 0.11 |
AC102953.6 |
|
2408 |
0.22 |
| chr22_30768667_30768959 | 0.11 |
RP1-130H16.16 |
|
5049 |
0.09 |
| chr6_42013197_42013548 | 0.11 |
CCND3 |
cyclin D3 |
3052 |
0.21 |
| chr19_3136242_3137376 | 0.11 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
618 |
0.58 |
| chr17_40441173_40441395 | 0.11 |
STAT5A |
signal transducer and activator of transcription 5A |
723 |
0.54 |
| chr22_50747133_50747284 | 0.11 |
PLXNB2 |
plexin B2 |
1152 |
0.29 |
| chr7_45016674_45016991 | 0.10 |
MYO1G |
myosin IG |
1865 |
0.25 |
| chr9_139000986_139001191 | 0.10 |
C9orf69 |
chromosome 9 open reading frame 69 |
9032 |
0.19 |
| chr19_12776607_12776758 | 0.10 |
MAN2B1 |
mannosidase, alpha, class 2B, member 1 |
866 |
0.26 |
| chrX_57313067_57313960 | 0.10 |
FAAH2 |
fatty acid amide hydrolase 2 |
374 |
0.92 |
| chr4_9154561_9154798 | 0.10 |
ENSG00000264372 |
. |
15341 |
0.12 |
| chr16_4714977_4715231 | 0.10 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
14703 |
0.11 |
| chr10_74080255_74080406 | 0.10 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
24389 |
0.15 |
| chr1_42192309_42192460 | 0.10 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
25712 |
0.21 |
| chr19_54677285_54677436 | 0.10 |
TMC4 |
transmembrane channel-like 4 |
416 |
0.64 |
| chr17_73567230_73567381 | 0.10 |
MYO15B |
myosin XVB pseudogene |
19553 |
0.1 |
| chr5_34930297_34930570 | 0.10 |
DNAJC21 |
DnaJ (Hsp40) homolog, subfamily C, member 21 |
735 |
0.64 |
| chr14_103577704_103577855 | 0.10 |
EXOC3L4 |
exocyst complex component 3-like 4 |
3926 |
0.19 |
| chr19_52132963_52133123 | 0.10 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
545 |
0.64 |
| chr15_40532500_40532651 | 0.10 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
148 |
0.93 |
| chr15_83348879_83349094 | 0.10 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
19006 |
0.12 |
| chr14_91839274_91839753 | 0.10 |
ENSG00000265856 |
. |
39456 |
0.15 |
| chr1_6085866_6086187 | 0.10 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
334 |
0.88 |
| chrX_106033404_106033689 | 0.10 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
12364 |
0.24 |
| chr19_42926797_42927063 | 0.09 |
LIPE |
lipase, hormone-sensitive |
425 |
0.63 |
| chr5_176785668_176785889 | 0.09 |
RGS14 |
regulator of G-protein signaling 14 |
940 |
0.38 |
| chr19_2041550_2041783 | 0.09 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
415 |
0.72 |
| chr19_3179462_3180177 | 0.09 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
1083 |
0.38 |
| chr17_76165715_76165934 | 0.09 |
SYNGR2 |
synaptogyrin 2 |
585 |
0.61 |
| chr9_92096324_92096488 | 0.09 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1601 |
0.45 |
| chr17_55339641_55339792 | 0.09 |
MSI2 |
musashi RNA-binding protein 2 |
5337 |
0.3 |
| chr17_56355369_56355598 | 0.09 |
MPO |
myeloperoxidase |
2813 |
0.18 |
| chr14_91731332_91731581 | 0.09 |
GPR68 |
G protein-coupled receptor 68 |
11187 |
0.16 |
| chr3_184098891_184099179 | 0.09 |
CHRD |
chordin |
928 |
0.39 |
| chrX_38922731_38922882 | 0.09 |
ENSG00000207122 |
. |
17732 |
0.3 |
| chr17_1548323_1548533 | 0.09 |
SCARF1 |
scavenger receptor class F, member 1 |
610 |
0.56 |
| chr9_116343099_116343250 | 0.09 |
RGS3 |
regulator of G-protein signaling 3 |
34 |
0.98 |
| chr1_6090319_6090470 | 0.09 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
3037 |
0.23 |
| chr12_113494960_113495179 | 0.09 |
DTX1 |
deltex homolog 1 (Drosophila) |
426 |
0.82 |
| chr7_45067326_45068016 | 0.09 |
CCM2 |
cerebral cavernous malformation 2 |
400 |
0.81 |
| chr18_72783416_72783729 | 0.09 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
133896 |
0.05 |
| chr6_35270906_35271095 | 0.09 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
5371 |
0.2 |
| chr20_58632411_58632562 | 0.09 |
C20orf197 |
chromosome 20 open reading frame 197 |
1506 |
0.49 |
| chr11_74699974_74700511 | 0.09 |
NEU3 |
sialidase 3 (membrane sialidase) |
252 |
0.9 |
| chr19_16696203_16696354 | 0.09 |
MED26 |
mediator complex subunit 26 |
2134 |
0.15 |
| chr16_85605071_85605483 | 0.09 |
GSE1 |
Gse1 coiled-coil protein |
39738 |
0.16 |
| chr7_63361181_63361806 | 0.09 |
ENSG00000263891 |
. |
47 |
0.98 |
| chr19_49977449_49977689 | 0.09 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
60 |
0.9 |
| chr9_34591301_34591802 | 0.09 |
CNTFR |
ciliary neurotrophic factor receptor |
1430 |
0.24 |
| chr9_95822109_95822464 | 0.09 |
SUSD3 |
sushi domain containing 3 |
1225 |
0.45 |
| chr10_81077160_81077479 | 0.09 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
11344 |
0.22 |
| chr1_17026830_17027051 | 0.09 |
ESPNP |
espin pseudogene |
10174 |
0.12 |
| chr1_40426265_40426496 | 0.09 |
MFSD2A |
major facilitator superfamily domain containing 2A |
5558 |
0.17 |
| chr8_103674687_103674993 | 0.09 |
KLF10 |
Kruppel-like factor 10 |
6710 |
0.25 |
| chr1_110158969_110159120 | 0.09 |
AMPD2 |
adenosine monophosphate deaminase 2 |
318 |
0.77 |
| chr3_32502059_32502315 | 0.09 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
42713 |
0.15 |
| chr19_52149205_52149440 | 0.09 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
524 |
0.49 |
| chrX_78621891_78622920 | 0.08 |
ITM2A |
integral membrane protein 2A |
451 |
0.91 |
| chr19_1840880_1841031 | 0.08 |
REXO1 |
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
7497 |
0.08 |
| chr2_242801310_242801461 | 0.08 |
PDCD1 |
programmed cell death 1 |
325 |
0.81 |
| chr1_175160611_175161799 | 0.08 |
KIAA0040 |
KIAA0040 |
685 |
0.78 |
| chr21_43639797_43640496 | 0.08 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
215 |
0.89 |
| chr1_41131794_41131945 | 0.08 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
540 |
0.74 |
| chr15_90754552_90754703 | 0.08 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
9859 |
0.12 |
| chr22_37639373_37639682 | 0.08 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
761 |
0.58 |
| chr17_46688443_46688779 | 0.08 |
HOXB7 |
homeobox B7 |
232 |
0.8 |
| chr22_37639776_37640249 | 0.08 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
276 |
0.88 |
| chrX_142723045_142723196 | 0.08 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
101 |
0.98 |
| chr2_235165280_235165431 | 0.08 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
205924 |
0.02 |
| chr1_28196256_28196434 | 0.08 |
THEMIS2 |
thymocyte selection associated family member 2 |
2710 |
0.16 |
| chr11_63974616_63975431 | 0.08 |
FERMT3 |
fermitin family member 3 |
266 |
0.78 |
| chr19_13206173_13206324 | 0.08 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
7433 |
0.1 |
| chrX_117662084_117662235 | 0.08 |
DOCK11 |
dedicator of cytokinesis 11 |
32287 |
0.18 |
| chr22_37881004_37881696 | 0.08 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
683 |
0.62 |
| chr10_5334923_5335219 | 0.08 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
4638 |
0.23 |
| chr3_129830565_129830801 | 0.08 |
FAM86HP |
family with sequence similarity 86, member H, pseudogene |
368 |
0.85 |
| chr2_145198575_145198734 | 0.08 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
10517 |
0.28 |
| chr2_113593127_113593420 | 0.08 |
IL1B |
interleukin 1, beta |
738 |
0.63 |
| chr17_4847711_4848500 | 0.08 |
RNF167 |
ring finger protein 167 |
1959 |
0.11 |
| chr3_52265762_52265913 | 0.08 |
TLR9 |
TLR9 |
631 |
0.53 |
| chr15_81590554_81590826 | 0.08 |
IL16 |
interleukin 16 |
1067 |
0.54 |
| chr3_5028227_5029021 | 0.08 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
6978 |
0.21 |
| chr9_74398314_74398465 | 0.08 |
TMEM2 |
transmembrane protein 2 |
14589 |
0.27 |
| chr20_31099035_31099434 | 0.08 |
C20orf112 |
chromosome 20 open reading frame 112 |
24 |
0.98 |
| chr17_35078057_35078719 | 0.08 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
119997 |
0.04 |
| chr5_76146227_76146557 | 0.08 |
S100Z |
S100 calcium binding protein Z |
468 |
0.79 |
| chr16_390161_390342 | 0.08 |
AXIN1 |
axin 1 |
12198 |
0.1 |
| chr11_67121279_67121678 | 0.08 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
461 |
0.64 |
| chr11_72493009_72493423 | 0.08 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
188 |
0.89 |
| chr5_131757138_131757303 | 0.08 |
AC116366.5 |
|
4926 |
0.14 |
| chr6_149805124_149805364 | 0.08 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
882 |
0.55 |
| chr9_215390_215630 | 0.08 |
DOCK8 |
dedicator of cytokinesis 8 |
339 |
0.52 |
| chr11_103947079_103947230 | 0.07 |
PDGFD |
platelet derived growth factor D |
39400 |
0.18 |
| chr11_65307798_65308115 | 0.07 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
33 |
0.95 |
| chr8_135700072_135700223 | 0.07 |
ZFAT |
zinc finger and AT hook domain containing |
8650 |
0.29 |
| chr9_129485287_129485658 | 0.07 |
ENSG00000266403 |
. |
4860 |
0.26 |
| chr12_133070120_133070271 | 0.07 |
FBRSL1 |
fibrosin-like 1 |
3038 |
0.27 |
| chr22_38576947_38577679 | 0.07 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
448 |
0.76 |
| chr11_44613932_44614083 | 0.07 |
CD82 |
CD82 molecule |
4399 |
0.23 |
| chr20_62199468_62200014 | 0.07 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
314 |
0.8 |
| chr14_103578008_103578159 | 0.07 |
EXOC3L4 |
exocyst complex component 3-like 4 |
4230 |
0.19 |
| chr6_166954866_166955076 | 0.07 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
967 |
0.56 |
| chr7_5595689_5595960 | 0.07 |
CTB-161C1.1 |
|
538 |
0.69 |
| chr10_74079018_74079438 | 0.07 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
25491 |
0.15 |
| chr1_21978657_21978808 | 0.07 |
RAP1GAP |
RAP1 GTPase activating protein |
384 |
0.87 |
| chr17_34128388_34128539 | 0.07 |
MMP28 |
matrix metallopeptidase 28 |
5752 |
0.1 |
| chr7_30517944_30518357 | 0.07 |
NOD1 |
nucleotide-binding oligomerization domain containing 1 |
102 |
0.97 |
| chr3_50267688_50267839 | 0.07 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
4039 |
0.13 |
| chr10_70982290_70982511 | 0.07 |
HKDC1 |
hexokinase domain containing 1 |
2341 |
0.24 |
| chr19_36233477_36233815 | 0.07 |
IGFLR1 |
IGF-like family receptor 1 |
292 |
0.71 |
| chr7_155386209_155386360 | 0.07 |
AC009403.2 |
Protein LOC100506302 |
50811 |
0.12 |
| chr11_67917703_67917854 | 0.07 |
CTD-2655K5.1 |
|
20513 |
0.14 |
| chr19_3145852_3146003 | 0.07 |
AC005264.2 |
|
9246 |
0.11 |
| chr4_15004964_15005278 | 0.07 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
401 |
0.91 |
| chr3_52140725_52140876 | 0.07 |
LINC00696 |
long intergenic non-protein coding RNA 696 |
43233 |
0.09 |
| chr19_43857990_43858141 | 0.07 |
CD177 |
CD177 molecule |
240 |
0.92 |
| chr2_7059033_7059408 | 0.07 |
ENSG00000228203 |
. |
380 |
0.82 |
| chr17_66340892_66341141 | 0.07 |
ARSG |
arylsulfatase G |
53357 |
0.11 |
| chr9_137235953_137236130 | 0.07 |
RXRA |
retinoid X receptor, alpha |
17615 |
0.22 |
| chr11_67045241_67045509 | 0.07 |
ANKRD13D |
ankyrin repeat domain 13 family, member D |
10643 |
0.11 |
| chr19_35985015_35985166 | 0.07 |
KRTDAP |
keratinocyte differentiation-associated protein |
3657 |
0.13 |
| chr5_1105065_1105874 | 0.07 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
6681 |
0.19 |
| chr15_31653239_31653495 | 0.07 |
KLF13 |
Kruppel-like factor 13 |
4990 |
0.34 |
| chr19_55836444_55836628 | 0.07 |
TMEM150B |
transmembrane protein 150B |
133 |
0.89 |
| chr3_15482302_15482608 | 0.07 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
354 |
0.48 |
| chr14_70161423_70161574 | 0.07 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
32119 |
0.19 |
| chr19_38539302_38539460 | 0.07 |
ENSG00000221258 |
. |
5224 |
0.2 |
| chr15_40627878_40628029 | 0.07 |
C15orf52 |
chromosome 15 open reading frame 52 |
2288 |
0.13 |
| chr20_58803254_58803405 | 0.07 |
ENSG00000207802 |
. |
80203 |
0.11 |
| chr10_45958891_45959042 | 0.07 |
RP11-67C2.2 |
|
10397 |
0.25 |
| chr17_29645805_29646348 | 0.07 |
CTD-2370N5.3 |
|
227 |
0.88 |
| chr18_13465441_13465744 | 0.07 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
578 |
0.59 |
| chr20_52195598_52195985 | 0.07 |
ZNF217 |
zinc finger protein 217 |
3225 |
0.23 |
| chr7_98871889_98872040 | 0.07 |
ARPC1A |
actin related protein 2/3 complex, subunit 1A, 41kDa |
51557 |
0.11 |
| chr2_221889944_221890095 | 0.07 |
EPHA4 |
EPH receptor A4 |
477261 |
0.01 |
| chr22_39885842_39885993 | 0.07 |
MIEF1 |
mitochondrial elongation factor 1 |
10188 |
0.13 |
| chr12_53718111_53718485 | 0.07 |
AAAS |
achalasia, adrenocortical insufficiency, alacrimia |
244 |
0.86 |
| chr13_41553204_41553401 | 0.07 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
3116 |
0.27 |
| chr1_92949289_92949440 | 0.07 |
GFI1 |
growth factor independent 1 transcription repressor |
147 |
0.97 |
| chr1_209943004_209943387 | 0.07 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
1235 |
0.37 |
| chr9_139923753_139923935 | 0.07 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
1104 |
0.19 |
| chr4_2789531_2789742 | 0.07 |
SH3BP2 |
SH3-domain binding protein 2 |
5114 |
0.22 |
| chr3_186598535_186598686 | 0.07 |
ADIPOQ-AS1 |
ADIPOQ antisense RNA 1 |
24698 |
0.12 |
| chr19_12660200_12660351 | 0.06 |
ZNF564 |
zinc finger protein 564 |
1973 |
0.19 |
| chr11_57333961_57334391 | 0.06 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
600 |
0.6 |
| chr9_95895594_95896005 | 0.06 |
NINJ1 |
ninjurin 1 |
771 |
0.63 |
| chr1_15279622_15279773 | 0.06 |
KAZN |
kazrin, periplakin interacting protein |
7282 |
0.31 |
| chr8_12294063_12294275 | 0.06 |
FAM86B2 |
family with sequence similarity 86, member B2 |
254 |
0.93 |
| chr16_11722953_11723104 | 0.06 |
LITAF |
lipopolysaccharide-induced TNF factor |
38 |
0.98 |
| chr11_68596728_68596879 | 0.06 |
CPT1A |
carnitine palmitoyltransferase 1A (liver) |
10349 |
0.2 |
| chr16_89427646_89427797 | 0.06 |
ANKRD11 |
ankyrin repeat domain 11 |
32327 |
0.11 |
| chr19_18113119_18113338 | 0.06 |
ARRDC2 |
arrestin domain containing 2 |
1287 |
0.37 |
| chr10_5295585_5295741 | 0.06 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
34770 |
0.13 |
| chr12_132899931_132900082 | 0.06 |
GALNT9 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
5783 |
0.19 |
| chr16_55533839_55533990 | 0.06 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
8996 |
0.21 |
| chr9_135937365_135937516 | 0.06 |
CEL |
carboxyl ester lipase |
75 |
0.96 |
| chr6_10643337_10643488 | 0.06 |
GCNT6 |
glucosaminyl (N-acetyl) transferase 6 |
9254 |
0.15 |
| chr19_16358640_16358791 | 0.06 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
20299 |
0.14 |
| chr18_28681584_28682412 | 0.06 |
DSC2 |
desmocollin 2 |
31 |
0.88 |
| chr8_144634447_144634598 | 0.06 |
GSDMD |
gasdermin D |
855 |
0.33 |
| chr11_73358513_73358664 | 0.06 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
6 |
0.98 |
| chr14_104048722_104048873 | 0.06 |
ENSG00000264904 |
. |
6401 |
0.09 |
| chr18_23670660_23670942 | 0.06 |
SS18 |
synovial sarcoma translocation, chromosome 18 |
64 |
0.98 |
| chr10_73516963_73517263 | 0.06 |
C10orf54 |
chromosome 10 open reading frame 54 |
274 |
0.92 |
| chr10_50395214_50395459 | 0.06 |
C10orf128 |
chromosome 10 open reading frame 128 |
1021 |
0.55 |
| chr6_43766546_43766697 | 0.06 |
VEGFA |
vascular endothelial growth factor A |
24531 |
0.16 |
| chr16_89790012_89790261 | 0.06 |
ZNF276 |
zinc finger protein 276 |
2184 |
0.16 |
| chr15_34875013_34876331 | 0.06 |
GOLGA8B |
golgin A8 family, member B |
99 |
0.96 |
| chr5_42991474_42991625 | 0.06 |
CTD-2035E11.3 |
|
26982 |
0.15 |
| chr16_27328627_27328778 | 0.06 |
IL4R |
interleukin 4 receptor |
3389 |
0.25 |
| chr3_184081214_184081580 | 0.06 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
175 |
0.89 |
| chr2_30372277_30372798 | 0.06 |
YPEL5 |
yippee-like 5 (Drosophila) |
2155 |
0.3 |
| chr8_66922647_66922798 | 0.06 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
11073 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |