Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
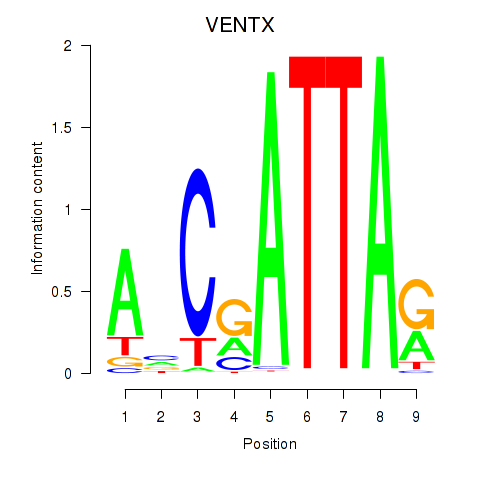
Results for VENTX
Z-value: 0.49
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENT homeobox |
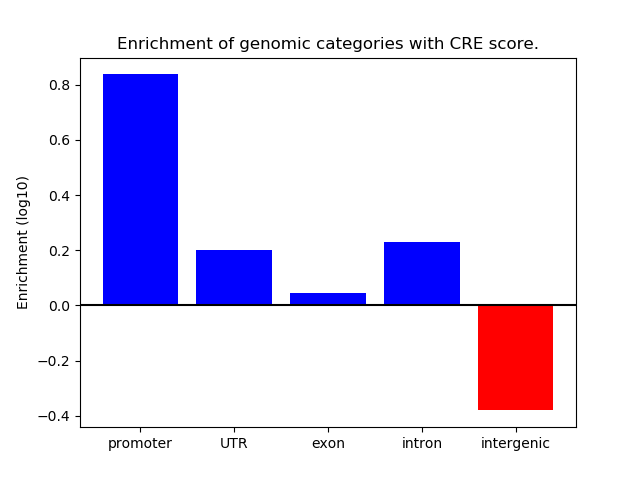
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_135050883_135051034 | VENTX | 50 | 0.956989 | 0.86 | 3.1e-03 | Click! |
| chr10_135050086_135050399 | VENTX | 666 | 0.567785 | 0.63 | 7.1e-02 | Click! |
| chr10_135055376_135055527 | VENTX | 4543 | 0.126368 | 0.51 | 1.6e-01 | Click! |
| chr10_135054758_135055166 | VENTX | 4054 | 0.132122 | -0.29 | 4.4e-01 | Click! |
| chr10_135050414_135050565 | VENTX | 419 | 0.744992 | 0.23 | 5.5e-01 | Click! |
Activity of the VENTX motif across conditions
Conditions sorted by the z-value of the VENTX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
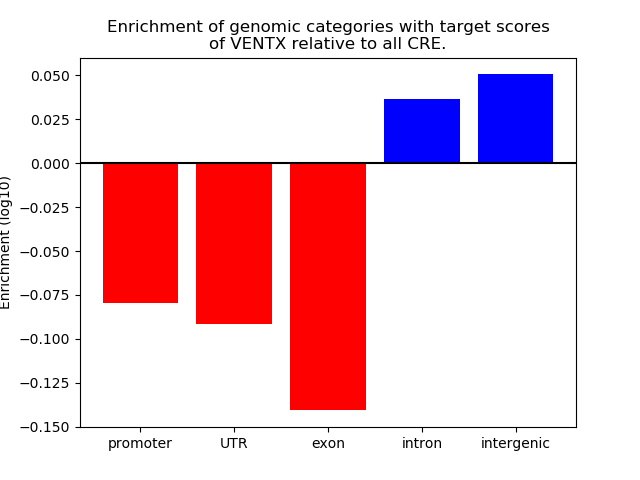
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_180523773_180523999 | 0.17 |
OR2V1 |
olfactory receptor, family 2, subfamily V, member 1 |
28418 |
0.12 |
| chr5_180529470_180529621 | 0.16 |
OR2V1 |
olfactory receptor, family 2, subfamily V, member 1 |
22759 |
0.13 |
| chr8_18870397_18870669 | 0.16 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
663 |
0.74 |
| chr1_210412971_210413268 | 0.14 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
5727 |
0.24 |
| chr10_62332469_62332822 | 0.13 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
224 |
0.97 |
| chr2_148394040_148394242 | 0.13 |
ENSG00000253083 |
. |
76982 |
0.1 |
| chr1_112604147_112604298 | 0.12 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
72445 |
0.11 |
| chr6_14671059_14671210 | 0.12 |
ENSG00000206960 |
. |
24368 |
0.28 |
| chr9_13278507_13279169 | 0.11 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr13_108520152_108520684 | 0.11 |
FAM155A |
family with sequence similarity 155, member A |
1335 |
0.55 |
| chr2_188418561_188419245 | 0.11 |
AC007319.1 |
|
27 |
0.69 |
| chr15_47476270_47477144 | 0.10 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr7_25901481_25901888 | 0.10 |
ENSG00000199085 |
. |
87922 |
0.1 |
| chr11_27743805_27744092 | 0.10 |
BDNF |
brain-derived neurotrophic factor |
343 |
0.92 |
| chr10_114714851_114715858 | 0.10 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr14_53275157_53275308 | 0.10 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
16846 |
0.16 |
| chr16_62068318_62068835 | 0.10 |
CDH8 |
cadherin 8, type 2 |
462 |
0.9 |
| chr18_53253101_53253291 | 0.10 |
TCF4 |
transcription factor 4 |
1 |
0.99 |
| chr3_148716623_148716961 | 0.09 |
GYG1 |
glycogenin 1 |
6634 |
0.21 |
| chr19_56332953_56333104 | 0.09 |
NLRP11 |
NLR family, pyrin domain containing 11 |
3425 |
0.16 |
| chr6_123110919_123111172 | 0.09 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
574 |
0.77 |
| chr17_59534634_59534785 | 0.09 |
TBX4 |
T-box 4 |
902 |
0.53 |
| chr6_133563559_133563958 | 0.09 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
989 |
0.7 |
| chr3_100118869_100119020 | 0.09 |
LNP1 |
leukemia NUP98 fusion partner 1 |
1093 |
0.41 |
| chr4_105982764_105983024 | 0.09 |
ENSG00000252136 |
. |
44250 |
0.17 |
| chr13_107124235_107124715 | 0.09 |
EFNB2 |
ephrin-B2 |
62987 |
0.14 |
| chr10_52750817_52751095 | 0.08 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
11 |
0.99 |
| chr11_57077918_57078147 | 0.08 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
11639 |
0.11 |
| chr1_30417491_30417642 | 0.08 |
ENSG00000222787 |
. |
59817 |
0.17 |
| chr11_10661867_10662018 | 0.08 |
MRVI1 |
murine retrovirus integration site 1 homolog |
11790 |
0.19 |
| chr11_57546160_57546525 | 0.08 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
2733 |
0.22 |
| chr10_96161850_96162001 | 0.08 |
TBC1D12 |
TBC1 domain family, member 12 |
336 |
0.91 |
| chr2_182760406_182760853 | 0.08 |
SSFA2 |
sperm specific antigen 2 |
3706 |
0.25 |
| chr20_59830411_59830643 | 0.08 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
3045 |
0.42 |
| chr10_126846286_126846503 | 0.08 |
CTBP2 |
C-terminal binding protein 2 |
891 |
0.72 |
| chr13_67804057_67805835 | 0.08 |
PCDH9 |
protocadherin 9 |
478 |
0.9 |
| chr18_33164100_33164377 | 0.08 |
GALNT1 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) |
2534 |
0.31 |
| chr19_1154555_1155129 | 0.08 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr1_214722890_214723041 | 0.08 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
1601 |
0.51 |
| chr8_126322078_126322527 | 0.08 |
ENSG00000242170 |
. |
39456 |
0.16 |
| chr8_42356526_42357223 | 0.08 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1887 |
0.34 |
| chr2_119989374_119989591 | 0.08 |
STEAP3 |
STEAP family member 3, metalloreductase |
8053 |
0.2 |
| chr4_57984994_57985389 | 0.08 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
8640 |
0.18 |
| chr22_41178472_41178623 | 0.08 |
SLC25A17 |
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
2349 |
0.27 |
| chr22_46476561_46476775 | 0.07 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr22_46471692_46472277 | 0.07 |
FLJ27365 |
hsa-mir-4763 |
4208 |
0.11 |
| chr14_34401158_34401309 | 0.07 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
18799 |
0.24 |
| chr9_39816869_39817104 | 0.07 |
SPATA31A2 |
SPATA31 subfamily A, member 2 |
67989 |
0.13 |
| chr17_12645469_12645648 | 0.07 |
MYOCD |
myocardin |
2110 |
0.29 |
| chr14_27066054_27066282 | 0.07 |
NOVA1 |
neuro-oncological ventral antigen 1 |
120 |
0.95 |
| chr10_29010987_29011704 | 0.07 |
ENSG00000201001 |
. |
38387 |
0.14 |
| chr9_39287616_39287852 | 0.07 |
CNTNAP3 |
contactin associated protein-like 3 |
358 |
0.92 |
| chr15_67334428_67334662 | 0.07 |
SMAD3 |
SMAD family member 3 |
21556 |
0.25 |
| chr9_27109543_27109972 | 0.07 |
TEK |
TEK tyrosine kinase, endothelial |
306 |
0.93 |
| chr1_8433877_8434267 | 0.07 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
9174 |
0.19 |
| chr5_139721479_139721638 | 0.07 |
HBEGF |
heparin-binding EGF-like growth factor |
4630 |
0.14 |
| chr4_77115816_77116180 | 0.07 |
SCARB2 |
scavenger receptor class B, member 2 |
18758 |
0.15 |
| chr5_16935133_16935934 | 0.07 |
MYO10 |
myosin X |
514 |
0.81 |
| chr9_22239273_22239447 | 0.07 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
125683 |
0.06 |
| chr21_27540114_27540265 | 0.07 |
APP |
amyloid beta (A4) precursor protein |
2783 |
0.28 |
| chr4_43019583_43019734 | 0.07 |
GRXCR1 |
glutaredoxin, cysteine rich 1 |
124374 |
0.06 |
| chr4_158143735_158144020 | 0.07 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
1059 |
0.6 |
| chr9_43685064_43685767 | 0.07 |
CNTNAP3B |
contactin associated protein-like 3B |
505 |
0.86 |
| chrX_40048176_40048340 | 0.07 |
BCOR |
BCL6 corepressor |
11676 |
0.29 |
| chr1_230223080_230223231 | 0.07 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20137 |
0.23 |
| chr12_32739224_32739514 | 0.07 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
12091 |
0.26 |
| chr4_83810146_83810528 | 0.07 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
1834 |
0.28 |
| chr1_95267376_95267764 | 0.07 |
SLC44A3 |
solute carrier family 44, member 3 |
18328 |
0.2 |
| chr6_42192493_42192832 | 0.07 |
MRPS10 |
mitochondrial ribosomal protein S10 |
7059 |
0.19 |
| chr2_9389435_9389677 | 0.07 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
42662 |
0.19 |
| chr21_16433048_16433989 | 0.07 |
NRIP1 |
nuclear receptor interacting protein 1 |
3608 |
0.34 |
| chr1_108479731_108479983 | 0.07 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
27208 |
0.19 |
| chr18_9016920_9017224 | 0.07 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
85556 |
0.08 |
| chr3_121450033_121450184 | 0.07 |
GOLGB1 |
golgin B1 |
1311 |
0.47 |
| chr18_59659824_59660127 | 0.07 |
RNF152 |
ring finger protein 152 |
98511 |
0.08 |
| chr14_23289433_23289584 | 0.07 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
488 |
0.61 |
| chr9_41432727_41433091 | 0.06 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
67770 |
0.13 |
| chr5_14184300_14184719 | 0.06 |
TRIO |
trio Rho guanine nucleotide exchange factor |
602 |
0.86 |
| chr1_95005421_95005705 | 0.06 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
1630 |
0.52 |
| chr4_88817282_88817552 | 0.06 |
MEPE |
matrix extracellular phosphoglycoprotein |
63278 |
0.1 |
| chr1_85960933_85961288 | 0.06 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
30283 |
0.18 |
| chr17_45197869_45198099 | 0.06 |
ENSG00000221016 |
. |
2116 |
0.25 |
| chr16_4900453_4900687 | 0.06 |
UBN1 |
ubinuclein 1 |
1646 |
0.27 |
| chr1_179110770_179111258 | 0.06 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
1165 |
0.52 |
| chr17_37786060_37786211 | 0.06 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
1384 |
0.29 |
| chr10_96163775_96164021 | 0.06 |
TBC1D12 |
TBC1 domain family, member 12 |
1637 |
0.43 |
| chr14_65170645_65170998 | 0.06 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
1 |
0.98 |
| chr4_139160727_139161292 | 0.06 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
2494 |
0.42 |
| chr6_101846988_101847139 | 0.06 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
91 |
0.99 |
| chr16_13401641_13401792 | 0.06 |
AC003009.1 |
|
24724 |
0.26 |
| chr9_40632604_40633916 | 0.06 |
SPATA31A3 |
SPATA31 subfamily A, member 3 |
67031 |
0.14 |
| chrX_112573298_112573449 | 0.06 |
ENSG00000238811 |
. |
329017 |
0.01 |
| chr9_110310779_110311295 | 0.06 |
KLF4 |
Kruppel-like factor 4 (gut) |
58274 |
0.13 |
| chr4_60684263_60684575 | 0.06 |
ENSG00000201676 |
. |
14474 |
0.24 |
| chr2_43134028_43134374 | 0.06 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
114469 |
0.06 |
| chr12_54369798_54369949 | 0.06 |
HOTAIR |
HOX transcript antisense RNA |
1133 |
0.21 |
| chr20_61049732_61049883 | 0.06 |
GATA5 |
GATA binding protein 5 |
1219 |
0.43 |
| chr8_94935365_94935516 | 0.06 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
3333 |
0.24 |
| chr9_21677527_21678485 | 0.06 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr2_185462344_185462621 | 0.06 |
ZNF804A |
zinc finger protein 804A |
611 |
0.86 |
| chr19_30792538_30792689 | 0.06 |
ZNF536 |
zinc finger protein 536 |
70706 |
0.12 |
| chr3_49447935_49448923 | 0.06 |
RHOA |
ras homolog family member A |
932 |
0.32 |
| chr18_42595960_42596214 | 0.06 |
ENSG00000265957 |
. |
45956 |
0.2 |
| chr1_98514169_98514606 | 0.06 |
ENSG00000225206 |
. |
2660 |
0.42 |
| chr4_129731963_129732326 | 0.06 |
JADE1 |
jade family PHD finger 1 |
299 |
0.94 |
| chr3_128914407_128914640 | 0.06 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
11758 |
0.14 |
| chr19_42712554_42712705 | 0.06 |
DEDD2 |
death effector domain containing 2 |
9192 |
0.09 |
| chr4_30726767_30727321 | 0.06 |
PCDH7 |
protocadherin 7 |
3067 |
0.38 |
| chr6_11795100_11795449 | 0.06 |
ADTRP |
androgen-dependent TFPI-regulating protein |
12005 |
0.28 |
| chr17_70119242_70119466 | 0.06 |
SOX9 |
SRY (sex determining region Y)-box 9 |
2193 |
0.46 |
| chr1_246034788_246034939 | 0.06 |
RP11-83A16.1 |
|
161990 |
0.04 |
| chr4_90410428_90410579 | 0.06 |
RP11-115D19.1 |
|
62004 |
0.15 |
| chr11_12132484_12132785 | 0.06 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
496 |
0.87 |
| chr18_24237635_24237786 | 0.06 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
345 |
0.86 |
| chr9_65576390_65577630 | 0.06 |
SPATA31A7 |
SPATA31 subfamily A, member 7 |
67400 |
0.15 |
| chrX_105065880_105066259 | 0.06 |
NRK |
Nik related kinase |
467 |
0.87 |
| chr15_101419100_101419461 | 0.06 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
301 |
0.92 |
| chr13_30415572_30415723 | 0.06 |
UBL3 |
ubiquitin-like 3 |
9174 |
0.3 |
| chr3_170527527_170527678 | 0.06 |
ENSG00000222411 |
. |
32359 |
0.19 |
| chr6_4370257_4370408 | 0.05 |
ENSG00000201185 |
. |
57865 |
0.15 |
| chr1_15587858_15588078 | 0.05 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
9687 |
0.21 |
| chr2_66663569_66663965 | 0.05 |
MEIS1 |
Meis homeobox 1 |
758 |
0.57 |
| chr14_29859562_29859789 | 0.05 |
ENSG00000257522 |
. |
11813 |
0.29 |
| chr9_89952337_89952906 | 0.05 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr2_182758331_182758678 | 0.05 |
SSFA2 |
sperm specific antigen 2 |
1581 |
0.37 |
| chr7_28725956_28726399 | 0.05 |
CREB5 |
cAMP responsive element binding protein 5 |
579 |
0.86 |
| chr12_18841597_18841748 | 0.05 |
PLCZ1 |
phospholipase C, zeta 1 |
6299 |
0.25 |
| chr12_89744161_89744444 | 0.05 |
DUSP6 |
dual specificity phosphatase 6 |
646 |
0.8 |
| chr10_4100667_4100818 | 0.05 |
KLF6 |
Kruppel-like factor 6 |
273269 |
0.02 |
| chr17_32571233_32571384 | 0.05 |
CCL2 |
chemokine (C-C motif) ligand 2 |
10996 |
0.13 |
| chr14_31022770_31022975 | 0.05 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
5457 |
0.25 |
| chr2_237073495_237073728 | 0.05 |
AC079135.1 |
|
2479 |
0.22 |
| chr1_46714502_46714653 | 0.05 |
RAD54L |
RAD54-like (S. cerevisiae) |
1136 |
0.42 |
| chr7_113054663_113054814 | 0.05 |
TSRM |
Uncharacterized protein; Zinc finger domain-related protein TSRM |
36389 |
0.24 |
| chr2_106593474_106593625 | 0.05 |
C2orf40 |
chromosome 2 open reading frame 40 |
86201 |
0.09 |
| chr22_50730996_50731325 | 0.05 |
PLXNB2 |
plexin B2 |
3170 |
0.12 |
| chrX_81845191_81845342 | 0.05 |
ENSG00000202183 |
. |
28616 |
0.27 |
| chr2_101620301_101620452 | 0.05 |
RPL31 |
ribosomal protein L31 |
1175 |
0.38 |
| chr9_105966807_105967305 | 0.05 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
209430 |
0.03 |
| chr10_8105207_8105358 | 0.05 |
GATA3 |
GATA binding protein 3 |
8513 |
0.32 |
| chr8_12976784_12976935 | 0.05 |
DLC1 |
deleted in liver cancer 1 |
3106 |
0.31 |
| chr1_180470891_180472111 | 0.05 |
ACBD6 |
acyl-CoA binding domain containing 6 |
588 |
0.75 |
| chr12_10868992_10869331 | 0.05 |
YBX3 |
Y box binding protein 3 |
6745 |
0.18 |
| chr13_92456795_92456946 | 0.05 |
ENSG00000252508 |
. |
255227 |
0.02 |
| chr14_70168154_70168305 | 0.05 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
25388 |
0.21 |
| chr5_98111445_98111873 | 0.05 |
RGMB |
repulsive guidance molecule family member b |
2320 |
0.3 |
| chr5_121517177_121517861 | 0.05 |
CTC-441N14.1 |
|
25809 |
0.17 |
| chr3_179168052_179168298 | 0.05 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
758 |
0.66 |
| chr18_59619337_59619488 | 0.05 |
RNF152 |
ring finger protein 152 |
57948 |
0.16 |
| chrX_133500265_133500416 | 0.05 |
PHF6 |
PHD finger protein 6 |
6943 |
0.21 |
| chr11_69454777_69455129 | 0.05 |
CCND1 |
cyclin D1 |
902 |
0.63 |
| chr1_155971887_155972038 | 0.05 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
4899 |
0.11 |
| chr1_217542715_217542866 | 0.05 |
RP11-361K17.2 |
|
122527 |
0.06 |
| chr8_17015787_17015938 | 0.05 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
919 |
0.67 |
| chr8_89289368_89289519 | 0.05 |
RP11-586K2.1 |
|
49622 |
0.16 |
| chr12_109901272_109901510 | 0.05 |
KCTD10 |
potassium channel tetramerization domain containing 10 |
3191 |
0.21 |
| chr9_80499174_80499388 | 0.05 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
61366 |
0.15 |
| chr7_54911574_54911725 | 0.05 |
ENSG00000252054 |
. |
21862 |
0.24 |
| chr3_100785098_100785499 | 0.05 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
72939 |
0.11 |
| chr2_8260069_8260430 | 0.05 |
ENSG00000221255 |
. |
543277 |
0.0 |
| chr5_147836900_147837051 | 0.05 |
CTD-2283N19.1 |
|
26605 |
0.18 |
| chr17_29818089_29818240 | 0.05 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
3038 |
0.19 |
| chr2_65009133_65009284 | 0.05 |
ENSG00000253082 |
. |
5970 |
0.22 |
| chr1_224840379_224840592 | 0.05 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
36490 |
0.15 |
| chr10_12109593_12109744 | 0.05 |
DHTKD1 |
dehydrogenase E1 and transketolase domain containing 1 |
1303 |
0.41 |
| chr2_161237855_161238493 | 0.05 |
ENSG00000252465 |
. |
15305 |
0.2 |
| chr12_42875750_42875901 | 0.05 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
1049 |
0.48 |
| chr22_19073564_19073715 | 0.05 |
AC004471.9 |
|
35403 |
0.09 |
| chr6_106830032_106830183 | 0.05 |
RP11-404H14.1 |
|
22515 |
0.15 |
| chr14_85998039_85998552 | 0.05 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
1723 |
0.42 |
| chr10_61624710_61624861 | 0.05 |
CCDC6 |
coiled-coil domain containing 6 |
41629 |
0.19 |
| chr4_117414362_117414513 | 0.05 |
MTRNR2L13 |
MT-RNR2-like 13 (pseudogene) |
194421 |
0.03 |
| chr6_108883421_108883945 | 0.05 |
FOXO3 |
forkhead box O3 |
1614 |
0.53 |
| chr11_72452051_72452202 | 0.05 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11322 |
0.12 |
| chr15_99449448_99449599 | 0.05 |
RP11-654A16.1 |
|
12759 |
0.21 |
| chr10_17256481_17257128 | 0.05 |
VIM-AS1 |
VIM antisense RNA 1 |
12093 |
0.15 |
| chr17_41668751_41668949 | 0.05 |
ETV4 |
ets variant 4 |
11862 |
0.16 |
| chr14_86047408_86047594 | 0.05 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
50929 |
0.16 |
| chr11_111810760_111811015 | 0.05 |
DIXDC1 |
DIX domain containing 1 |
2736 |
0.15 |
| chr2_197089791_197089942 | 0.05 |
ENSG00000239161 |
. |
9820 |
0.19 |
| chr7_12728576_12729094 | 0.05 |
ARL4A |
ADP-ribosylation factor-like 4A |
1575 |
0.44 |
| chr11_131847622_131847821 | 0.05 |
RP11-697E14.2 |
|
6834 |
0.28 |
| chr6_126890888_126891039 | 0.05 |
ENSG00000201613 |
. |
20576 |
0.27 |
| chr16_79693058_79693209 | 0.05 |
ENSG00000221330 |
. |
10501 |
0.29 |
| chr5_88178054_88178448 | 0.05 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr8_10587151_10587560 | 0.05 |
CTD-2135J3.3 |
|
267 |
0.76 |
| chr3_114790766_114791082 | 0.05 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
702 |
0.82 |
| chr6_168079333_168079613 | 0.05 |
AL009178.1 |
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
116737 |
0.06 |
| chr5_178202811_178202962 | 0.05 |
AACSP1 |
acetoacetyl-CoA synthetase pseudogene 1 |
21726 |
0.18 |
| chr12_58705170_58705321 | 0.05 |
RP11-362K2.2 |
Protein LOC100506869 |
232662 |
0.02 |
| chr1_200558260_200558411 | 0.05 |
KIF14 |
kinesin family member 14 |
31527 |
0.2 |
| chr3_69870376_69870665 | 0.05 |
MITF |
microphthalmia-associated transcription factor |
44843 |
0.17 |
| chr11_63775179_63775448 | 0.05 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
20999 |
0.1 |
| chr1_226220312_226220463 | 0.05 |
H3F3A |
H3 histone, family 3A |
29165 |
0.13 |
| chr20_10521227_10521501 | 0.05 |
SLX4IP |
SLX4 interacting protein |
105413 |
0.06 |
| chr5_95584344_95584495 | 0.05 |
ENSG00000206997 |
. |
38488 |
0.2 |
| chr6_7547044_7547195 | 0.05 |
DSP |
desmoplakin |
5269 |
0.22 |
| chr6_136570446_136570737 | 0.05 |
MTFR2 |
mitochondrial fission regulator 2 |
841 |
0.63 |
| chr4_157880787_157880981 | 0.05 |
PDGFC |
platelet derived growth factor C |
11171 |
0.22 |
| chr5_81064717_81065068 | 0.05 |
SSBP2 |
single-stranded DNA binding protein 2 |
17820 |
0.28 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |