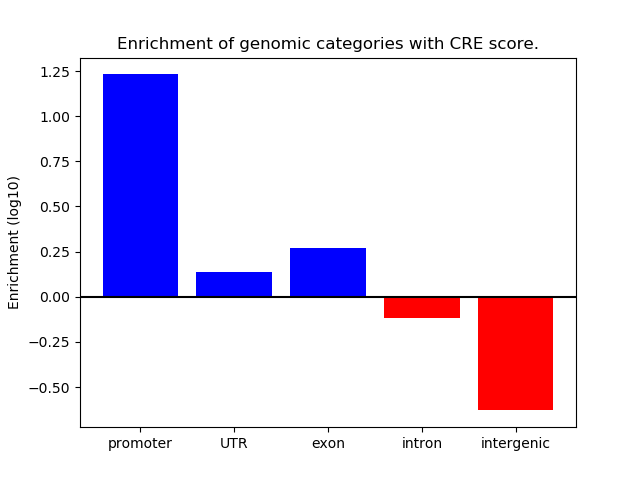
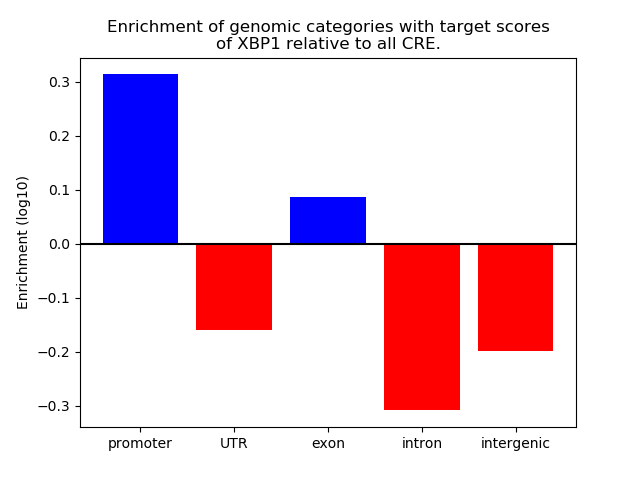
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
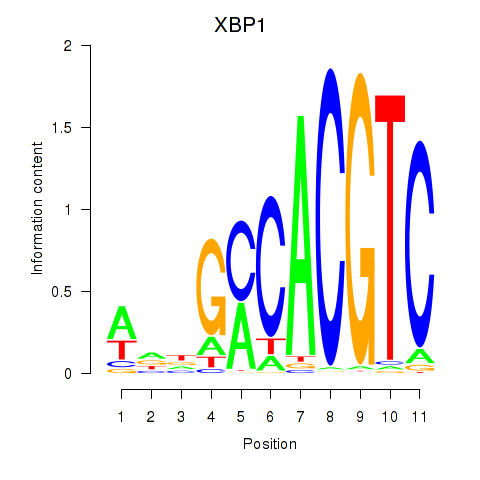
Results for XBP1
Z-value: 1.58
Transcription factors associated with XBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
XBP1
|
ENSG00000100219.12 | X-box binding protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_29191705_29191856 | XBP1 | 4341 | 0.154178 | -0.67 | 4.9e-02 | Click! |
| chr22_29195834_29196065 | XBP1 | 172 | 0.855072 | 0.60 | 9.0e-02 | Click! |
| chr22_29192759_29192910 | XBP1 | 3287 | 0.170616 | -0.49 | 1.8e-01 | Click! |
| chr22_29194035_29194328 | XBP1 | 1940 | 0.231288 | 0.38 | 3.2e-01 | Click! |
| chr22_29194979_29195773 | XBP1 | 745 | 0.488947 | 0.38 | 3.2e-01 | Click! |
Activity of the XBP1 motif across conditions
Conditions sorted by the z-value of the XBP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
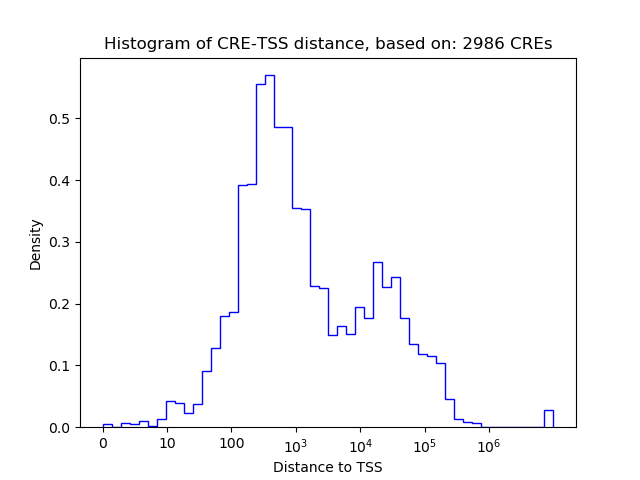
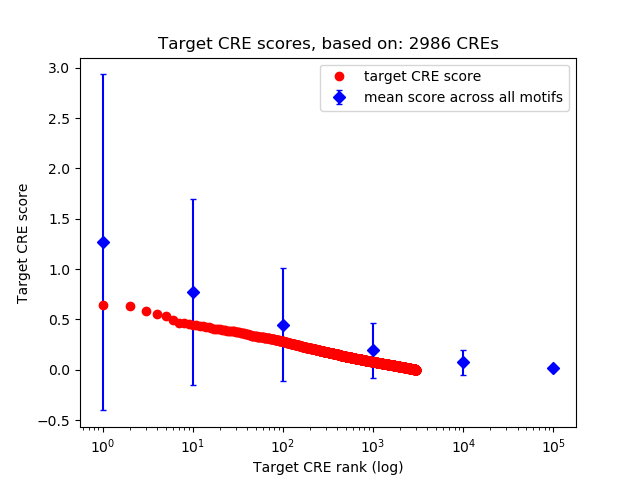
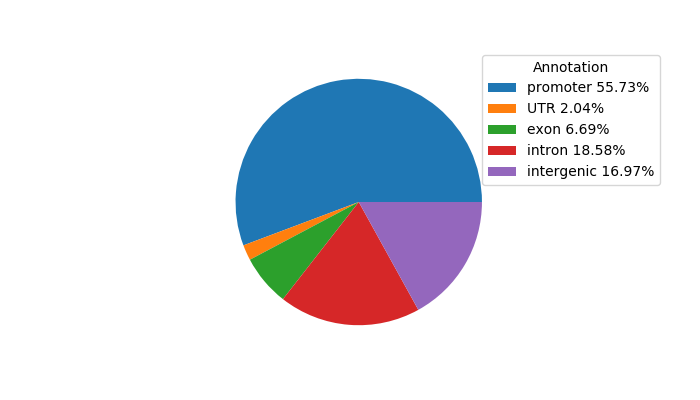
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_34184837_34185306 | 0.64 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
207115 |
0.02 |
| chr20_57467710_57467928 | 0.64 |
GNAS |
GNAS complex locus |
431 |
0.77 |
| chrX_134479032_134479223 | 0.59 |
ZNF449 |
zinc finger protein 449 |
406 |
0.77 |
| chr2_220408420_220408618 | 0.56 |
CHPF |
chondroitin polymerizing factor |
10 |
0.62 |
| chr3_18486507_18486658 | 0.54 |
ENSG00000228956 |
. |
66 |
0.7 |
| chr15_67361022_67361173 | 0.49 |
SMAD3 |
SMAD family member 3 |
2914 |
0.37 |
| chr14_76446267_76446546 | 0.47 |
TGFB3 |
transforming growth factor, beta 3 |
930 |
0.62 |
| chr21_40406107_40406258 | 0.46 |
ENSG00000272015 |
. |
139473 |
0.04 |
| chrX_39680179_39680466 | 0.46 |
ENSG00000263972 |
. |
16493 |
0.24 |
| chr16_65157518_65157704 | 0.45 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
1343 |
0.63 |
| chr3_9791200_9791462 | 0.45 |
OGG1 |
8-oxoguanine DNA glycosylase |
297 |
0.82 |
| chr16_89830973_89831469 | 0.44 |
FANCA |
Fanconi anemia, complementation group A |
19751 |
0.11 |
| chr3_134204282_134204486 | 0.43 |
CEP63 |
centrosomal protein 63kDa |
201 |
0.75 |
| chrX_108780083_108780293 | 0.43 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
83 |
0.98 |
| chr19_58459192_58459462 | 0.43 |
ZNF256 |
zinc finger protein 256 |
284 |
0.84 |
| chr3_196044847_196045072 | 0.42 |
TCTEX1D2 |
Tctex1 domain containing 2 |
200 |
0.53 |
| chr4_171030941_171031128 | 0.41 |
AADAT |
aminoadipate aminotransferase |
18184 |
0.26 |
| chr19_50031295_50031537 | 0.41 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
131 |
0.88 |
| chr10_45719589_45719755 | 0.40 |
ENSG00000207071 |
. |
35328 |
0.17 |
| chr19_3347090_3347241 | 0.40 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
12396 |
0.18 |
| chr2_238638558_238638814 | 0.40 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
37704 |
0.16 |
| chr20_48553311_48553551 | 0.40 |
RNF114 |
ring finger protein 114 |
483 |
0.72 |
| chr2_225792696_225792947 | 0.39 |
DOCK10 |
dedicator of cytokinesis 10 |
18961 |
0.28 |
| chr3_107243761_107243927 | 0.39 |
BBX |
bobby sox homolog (Drosophila) |
394 |
0.92 |
| chr12_4714049_4714390 | 0.38 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
68 |
0.97 |
| chr14_22996017_22996172 | 0.38 |
TRAJ15 |
T cell receptor alpha joining 15 |
2486 |
0.17 |
| chr1_12235343_12235565 | 0.38 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
8394 |
0.16 |
| chr8_29957431_29957780 | 0.38 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
1851 |
0.28 |
| chr1_92350650_92350871 | 0.38 |
TGFBR3 |
transforming growth factor, beta receptor III |
906 |
0.63 |
| chr19_8456957_8457108 | 0.37 |
RAB11B-AS1 |
RAB11B antisense RNA 1 |
1463 |
0.2 |
| chr10_76994278_76994482 | 0.37 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
1303 |
0.47 |
| chr1_203734376_203734572 | 0.37 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
169 |
0.94 |
| chr11_60691707_60691884 | 0.37 |
TMEM132A |
transmembrane protein 132A |
147 |
0.87 |
| chr15_60690387_60690946 | 0.37 |
ANXA2 |
annexin A2 |
275 |
0.94 |
| chr3_73672493_73672644 | 0.37 |
PDZRN3-AS1 |
PDZRN3 antisense RNA 1 |
151 |
0.94 |
| chr1_109633400_109633594 | 0.36 |
RP5-1065J22.8 |
|
17 |
0.94 |
| chr3_156271681_156272482 | 0.36 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
167 |
0.96 |
| chr15_86163319_86163521 | 0.36 |
RP11-815J21.3 |
|
7500 |
0.15 |
| chr1_67772983_67773214 | 0.36 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
51 |
0.98 |
| chr20_57426529_57426700 | 0.35 |
GNAS-AS1 |
GNAS antisense RNA 1 |
656 |
0.52 |
| chr11_61584736_61584906 | 0.35 |
FADS1 |
fatty acid desaturase 1 |
152 |
0.86 |
| chr14_23834178_23834353 | 0.35 |
EFS |
embryonal Fyn-associated substrate |
154 |
0.88 |
| chr19_20011402_20011620 | 0.35 |
ZNF93 |
zinc finger protein 93 |
226 |
0.9 |
| chr2_43038576_43038727 | 0.34 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
18919 |
0.23 |
| chr7_15722330_15722494 | 0.34 |
MEOX2 |
mesenchyme homeobox 2 |
4025 |
0.29 |
| chr10_44341794_44342095 | 0.34 |
ENSG00000238957 |
. |
188044 |
0.03 |
| chr20_37063439_37063660 | 0.33 |
ENSG00000200354 |
. |
908 |
0.36 |
| chr1_43736101_43736371 | 0.33 |
TMEM125 |
transmembrane protein 125 |
381 |
0.81 |
| chr3_52566513_52566726 | 0.33 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
928 |
0.41 |
| chr1_147860499_147860650 | 0.33 |
ENSG00000202064 |
. |
229 |
0.89 |
| chr1_214280862_214281029 | 0.33 |
RP11-53A1.2 |
|
76836 |
0.11 |
| chr19_54058906_54059057 | 0.33 |
ZNF331 |
zinc finger protein 331 |
240 |
0.86 |
| chr11_122050907_122051108 | 0.33 |
ENSG00000207994 |
. |
27991 |
0.16 |
| chr22_40271331_40271542 | 0.33 |
ENSG00000238875 |
. |
917 |
0.58 |
| chr2_182757184_182757414 | 0.33 |
SSFA2 |
sperm specific antigen 2 |
376 |
0.76 |
| chrX_47517975_47518127 | 0.33 |
UXT-AS1 |
UXT antisense RNA 1 |
181 |
0.76 |
| chr1_204485821_204486049 | 0.33 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
316 |
0.89 |
| chr17_15059171_15059366 | 0.33 |
ENSG00000238806 |
. |
81932 |
0.08 |
| chr15_65186326_65186539 | 0.33 |
ENSG00000264929 |
. |
6006 |
0.16 |
| chr19_20262264_20262447 | 0.32 |
CTC-260E6.9 |
|
362 |
0.76 |
| chr1_196847231_196847470 | 0.32 |
CFHR4 |
complement factor H-related 4 |
9794 |
0.22 |
| chr2_119602557_119602736 | 0.32 |
EN1 |
engrailed homeobox 1 |
2608 |
0.39 |
| chr9_136202256_136202547 | 0.32 |
SURF6 |
surfeit 6 |
834 |
0.32 |
| chr14_45366470_45366675 | 0.32 |
C14orf28 |
chromosome 14 open reading frame 28 |
34 |
0.8 |
| chr16_31084353_31084557 | 0.32 |
ZNF668 |
zinc finger protein 668 |
307 |
0.69 |
| chr8_38323279_38323475 | 0.32 |
FGFR1 |
fibroblast growth factor receptor 1 |
804 |
0.6 |
| chr2_25744901_25745083 | 0.32 |
AC104699.1 |
|
101006 |
0.07 |
| chr5_108745860_108746027 | 0.32 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
248 |
0.96 |
| chr6_39901443_39901850 | 0.32 |
MOCS1 |
molybdenum cofactor synthesis 1 |
514 |
0.84 |
| chr12_133658056_133658218 | 0.31 |
ZNF140 |
zinc finger protein 140 |
34 |
0.96 |
| chr13_102569247_102569398 | 0.31 |
FGF14 |
fibroblast growth factor 14 |
327 |
0.92 |
| chr19_42388528_42389094 | 0.31 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
296 |
0.84 |
| chr19_16438433_16439148 | 0.31 |
KLF2 |
Kruppel-like factor 2 |
3139 |
0.19 |
| chr13_40397646_40397797 | 0.31 |
ENSG00000212553 |
. |
33643 |
0.19 |
| chr9_133454530_133454782 | 0.31 |
FUBP3 |
far upstream element (FUSE) binding protein 3 |
337 |
0.91 |
| chr13_23754987_23755138 | 0.31 |
SGCG |
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
29 |
0.98 |
| chr2_62444590_62444741 | 0.30 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
2012 |
0.3 |
| chr17_21190411_21190564 | 0.30 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
861 |
0.6 |
| chr13_47216013_47216251 | 0.30 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
37839 |
0.21 |
| chr19_1026324_1026500 | 0.30 |
CNN2 |
calponin 2 |
114 |
0.9 |
| chr11_9634927_9635249 | 0.30 |
WEE1 |
WEE1 G2 checkpoint kinase |
26827 |
0.13 |
| chr2_122530051_122530202 | 0.30 |
TSN |
translin |
16379 |
0.21 |
| chr14_106967068_106967219 | 0.30 |
IGHV1-46 |
immunoglobulin heavy variable 1-46 |
645 |
0.33 |
| chrX_53110080_53110473 | 0.30 |
TSPYL2 |
TSPY-like 2 |
1273 |
0.44 |
| chr7_65235868_65236176 | 0.29 |
ENSG00000206785 |
. |
10983 |
0.15 |
| chr16_15247007_15247389 | 0.29 |
PKD1P6 |
polycystic kidney disease 1 (autosomal dominant) pseudogene 6 |
1223 |
0.28 |
| chr18_59221687_59221979 | 0.29 |
CDH20 |
cadherin 20, type 2 |
4734 |
0.36 |
| chr14_50087068_50087296 | 0.29 |
RPL36AL |
ribosomal protein L36a-like |
221 |
0.56 |
| chr5_119800069_119800772 | 0.29 |
PRR16 |
proline rich 16 |
401 |
0.92 |
| chr9_137288070_137288221 | 0.29 |
RXRA |
retinoid X receptor, alpha |
10283 |
0.23 |
| chr10_30959735_30960082 | 0.29 |
SVILP1 |
supervillin pseudogene 1 |
587 |
0.81 |
| chr20_22559380_22559531 | 0.29 |
FOXA2 |
forkhead box A2 |
5646 |
0.34 |
| chr1_23749079_23749230 | 0.29 |
TCEA3 |
transcription elongation factor A (SII), 3 |
2046 |
0.27 |
| chr17_18061433_18061699 | 0.29 |
MYO15A |
myosin XVA |
56 |
0.96 |
| chr3_72938425_72938589 | 0.29 |
GXYLT2 |
glucoside xylosyltransferase 2 |
1283 |
0.51 |
| chr15_86162061_86162404 | 0.29 |
RP11-815J21.3 |
|
8688 |
0.15 |
| chrX_65857892_65858837 | 0.29 |
EDA2R |
ectodysplasin A2 receptor |
519 |
0.86 |
| chr18_14852187_14852399 | 0.29 |
ENSG00000265499 |
. |
22128 |
0.19 |
| chr10_28034049_28034242 | 0.28 |
MKX |
mohawk homeobox |
378 |
0.52 |
| chr3_136153492_136153643 | 0.28 |
ENSG00000200571 |
. |
4542 |
0.28 |
| chr14_33366072_33366949 | 0.28 |
NPAS3 |
neuronal PAS domain protein 3 |
37629 |
0.23 |
| chr20_10903511_10903679 | 0.28 |
RP11-103J8.1 |
|
20663 |
0.27 |
| chr6_139308621_139308842 | 0.28 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
46 |
0.98 |
| chr14_71373640_71373972 | 0.28 |
PCNX |
pecanex homolog (Drosophila) |
316 |
0.94 |
| chr12_120218607_120218768 | 0.28 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
22500 |
0.2 |
| chr5_115177794_115178021 | 0.28 |
ATG12 |
autophagy related 12 |
352 |
0.71 |
| chrX_57932869_57933095 | 0.28 |
ZXDA |
zinc finger, X-linked, duplicated A |
4085 |
0.38 |
| chr2_237982035_237982186 | 0.27 |
COPS8 |
COP9 signalosome subunit 8 |
11845 |
0.18 |
| chrY_15018181_15018332 | 0.27 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
607 |
0.83 |
| chr3_196255461_196255682 | 0.27 |
SMCO1 |
single-pass membrane protein with coiled-coil domains 1 |
13334 |
0.12 |
| chr9_139972217_139972455 | 0.27 |
UAP1L1 |
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
257 |
0.74 |
| chr16_17107310_17107606 | 0.27 |
CTD-2576D5.4 |
|
120903 |
0.07 |
| chr15_45880472_45880722 | 0.27 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
574 |
0.56 |
| chr7_94023741_94023892 | 0.27 |
COL1A2 |
collagen, type I, alpha 2 |
57 |
0.99 |
| chr9_91309829_91309980 | 0.27 |
ENSG00000265873 |
. |
50916 |
0.18 |
| chr1_36107852_36108078 | 0.27 |
PSMB2 |
proteasome (prosome, macropain) subunit, beta type, 2 |
520 |
0.77 |
| chrX_800237_800388 | 0.26 |
SHOX |
short stature homeobox |
208770 |
0.02 |
| chr2_102803614_102803903 | 0.26 |
IL1RL2 |
interleukin 1 receptor-like 2 |
16 |
0.98 |
| chrY_750238_750389 | 0.26 |
NA |
NA |
> 106 |
NA |
| chr4_84030444_84030764 | 0.26 |
PLAC8 |
placenta-specific 8 |
392 |
0.89 |
| chr10_112116910_112117211 | 0.26 |
SMNDC1 |
survival motor neuron domain containing 1 |
52351 |
0.13 |
| chr1_155145539_155145727 | 0.26 |
KRTCAP2 |
keratinocyte associated protein 2 |
176 |
0.54 |
| chr4_39459578_39459788 | 0.26 |
RPL9 |
ribosomal protein L9 |
381 |
0.7 |
| chr2_174889838_174890106 | 0.26 |
SP3 |
Sp3 transcription factor |
59542 |
0.15 |
| chr1_239880855_239881135 | 0.26 |
ENSG00000233355 |
. |
1371 |
0.43 |
| chr1_89457123_89457381 | 0.26 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
1035 |
0.37 |
| chr2_237881760_237881911 | 0.26 |
ENSG00000202341 |
. |
42113 |
0.17 |
| chr2_99952225_99952767 | 0.26 |
TXNDC9 |
thioredoxin domain containing 9 |
300 |
0.84 |
| chr12_54746460_54746646 | 0.26 |
RP11-753H16.3 |
|
892 |
0.29 |
| chr2_228029012_228029295 | 0.26 |
COL4A3 |
collagen, type IV, alpha 3 (Goodpasture antigen) |
128 |
0.75 |
| chr9_115096315_115096686 | 0.26 |
PTBP3 |
polypyrimidine tract binding protein 3 |
553 |
0.78 |
| chr5_60458526_60459015 | 0.25 |
SMIM15 |
small integral membrane protein 15 |
469 |
0.67 |
| chr17_66592508_66592659 | 0.25 |
FAM20A |
family with sequence similarity 20, member A |
4947 |
0.26 |
| chr4_157115087_157115238 | 0.25 |
ENSG00000221189 |
. |
102636 |
0.08 |
| chr19_29703517_29704012 | 0.25 |
UQCRFS1 |
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
684 |
0.77 |
| chr22_18632897_18633158 | 0.25 |
USP18 |
ubiquitin specific peptidase 18 |
361 |
0.86 |
| chr16_2184293_2184536 | 0.25 |
ENSG00000265867 |
. |
1294 |
0.14 |
| chr8_11057352_11057503 | 0.25 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
1421 |
0.4 |
| chr1_149831891_149832049 | 0.25 |
HIST2H4B |
histone cluster 2, H4b |
734 |
0.36 |
| chr10_18468774_18468925 | 0.25 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
38740 |
0.2 |
| chr1_98510499_98510650 | 0.25 |
ENSG00000225206 |
. |
333 |
0.94 |
| chr6_19845920_19846071 | 0.25 |
RP1-167F1.2 |
|
6684 |
0.25 |
| chr12_12872464_12872615 | 0.25 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
1744 |
0.27 |
| chr15_92576579_92576730 | 0.24 |
RP11-24J19.1 |
|
139012 |
0.05 |
| chr6_157694117_157694268 | 0.24 |
ENSG00000252609 |
. |
18239 |
0.24 |
| chr9_73026328_73026548 | 0.24 |
KLF9 |
Kruppel-like factor 9 |
3102 |
0.35 |
| chr19_54962349_54962752 | 0.24 |
LENG8 |
leukocyte receptor cluster (LRC) member 8 |
83 |
0.93 |
| chr17_37123877_37124071 | 0.24 |
FBXO47 |
F-box protein 47 |
319 |
0.86 |
| chr16_15006581_15006965 | 0.24 |
RP11-958N24.1 |
|
1365 |
0.22 |
| chr6_170403243_170403627 | 0.24 |
RP11-302L19.1 |
|
74306 |
0.11 |
| chr3_128338529_128338709 | 0.24 |
RPN1 |
ribophorin I |
31073 |
0.16 |
| chr6_27100827_27101032 | 0.24 |
HIST1H2AG |
histone cluster 1, H2ag |
97 |
0.8 |
| chr2_239195506_239195657 | 0.24 |
PER2 |
period circadian clock 2 |
1626 |
0.26 |
| chr17_79010323_79010587 | 0.24 |
BAIAP2 |
BAI1-associated protein 2 |
652 |
0.61 |
| chrX_800923_801074 | 0.24 |
SHOX |
short stature homeobox |
209456 |
0.02 |
| chrY_750926_751077 | 0.24 |
NA |
NA |
> 106 |
NA |
| chr5_141443601_141443885 | 0.24 |
NDFIP1 |
Nedd4 family interacting protein 1 |
44327 |
0.13 |
| chr14_76044289_76044527 | 0.24 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
552 |
0.67 |
| chr7_69946308_69946459 | 0.24 |
AUTS2 |
autism susceptibility candidate 2 |
247742 |
0.02 |
| chr1_149804899_149805051 | 0.24 |
HIST2H4A |
histone cluster 2, H4a |
748 |
0.29 |
| chr9_101983648_101983918 | 0.24 |
ALG2 |
ALG2, alpha-1,3/1,6-mannosyltransferase |
454 |
0.53 |
| chr19_13983519_13983792 | 0.23 |
ENSG00000207613 |
. |
1858 |
0.15 |
| chr4_183004903_183005054 | 0.23 |
AC108142.1 |
|
1259 |
0.55 |
| chr13_102060461_102060612 | 0.23 |
NALCN |
sodium leak channel, non-selective |
8170 |
0.28 |
| chr14_100259771_100259922 | 0.23 |
EML1 |
echinoderm microtubule associated protein like 1 |
74 |
0.98 |
| chr10_14050350_14050950 | 0.23 |
FRMD4A |
FERM domain containing 4A |
118 |
0.97 |
| chr1_155715028_155715538 | 0.23 |
DAP3 |
death associated protein 3 |
8480 |
0.17 |
| chr14_61747147_61747473 | 0.23 |
PRKCH |
protein kinase C, eta |
361 |
0.67 |
| chr17_79818168_79818378 | 0.23 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
81 |
0.91 |
| chr22_45831450_45831605 | 0.23 |
RP1-102D24.5 |
|
13097 |
0.17 |
| chr21_36935983_36936169 | 0.23 |
ENSG00000211590 |
. |
156937 |
0.04 |
| chr20_4666407_4666914 | 0.23 |
PRNP |
prion protein |
222 |
0.95 |
| chr20_33680038_33680258 | 0.23 |
TRPC4AP |
transient receptor potential cation channel, subfamily C, member 4 associated protein |
462 |
0.78 |
| chr14_91225012_91225184 | 0.23 |
TTC7B |
tetratricopeptide repeat domain 7B |
27483 |
0.2 |
| chr16_16405241_16405629 | 0.23 |
ENSG00000265373 |
. |
1703 |
0.2 |
| chr18_29963098_29963249 | 0.22 |
GAREM |
GRB2 associated, regulator of MAPK1 |
87222 |
0.08 |
| chr7_128378913_128379302 | 0.22 |
CALU |
calumenin |
239 |
0.88 |
| chr5_175085682_175085863 | 0.22 |
HRH2 |
histamine receptor H2 |
739 |
0.7 |
| chr6_13486899_13487420 | 0.22 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
628 |
0.33 |
| chr1_205498395_205498557 | 0.22 |
CDK18 |
cyclin-dependent kinase 18 |
24546 |
0.13 |
| chr2_85132966_85133287 | 0.22 |
TMSB10 |
thymosin beta 10 |
377 |
0.86 |
| chr13_99506197_99506352 | 0.22 |
DOCK9 |
dedicator of cytokinesis 9 |
1964 |
0.38 |
| chr5_140090338_140090717 | 0.22 |
ENSG00000199990 |
. |
333 |
0.72 |
| chr10_133776022_133776178 | 0.22 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
19327 |
0.21 |
| chr16_74777769_74777965 | 0.22 |
FA2H |
fatty acid 2-hydroxylase |
2700 |
0.25 |
| chr15_90643522_90643980 | 0.22 |
IDH2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
176 |
0.93 |
| chrY_1318048_1318199 | 0.22 |
NA |
NA |
> 106 |
NA |
| chr20_34250346_34250754 | 0.22 |
CPNE1 |
copine I |
1930 |
0.18 |
| chr1_11753464_11753615 | 0.22 |
DRAXIN |
dorsal inhibitory axon guidance protein |
1753 |
0.22 |
| chr3_156272520_156272859 | 0.22 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
55 |
0.98 |
| chr9_136568137_136568308 | 0.22 |
SARDH |
sarcosine dehydrogenase |
601 |
0.75 |
| chr21_37528132_37528402 | 0.22 |
ENSG00000236830 |
. |
299 |
0.75 |
| chr21_44781922_44782352 | 0.22 |
SIK1 |
salt-inducible kinase 1 |
64871 |
0.13 |
| chr15_81589062_81589213 | 0.22 |
IL16 |
interleukin 16 |
117 |
0.97 |
| chr7_38351039_38351457 | 0.22 |
STARD3NL |
STARD3 N-terminal like |
133251 |
0.05 |
| chr5_178770702_178770853 | 0.22 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
1654 |
0.49 |
| chr6_27806026_27806177 | 0.21 |
HIST1H2AK |
histone cluster 1, H2ak |
16 |
0.77 |
| chrX_100872700_100873033 | 0.21 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
125 |
0.93 |
| chr11_35546514_35546852 | 0.21 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
468 |
0.87 |
| chr18_45968069_45968289 | 0.21 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
31056 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.3 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.1 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0071371 | cellular response to gonadotropin stimulus(GO:0071371) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.3 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0021612 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.2 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.1 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0042921 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.2 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:1903306 | negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0016241 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0050935 | sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:1902591 | vesicle coating(GO:0006901) single-organism membrane budding(GO:1902591) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.0 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.0 | 0.0 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.3 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.2 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.8 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |