Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
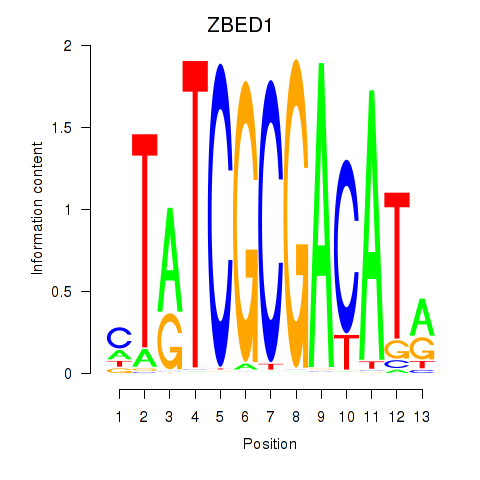
Results for ZBED1
Z-value: 0.28
Transcription factors associated with ZBED1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBED1
|
ENSG00000214717.5 | zinc finger BED-type containing 1 |
|
ZBED1
|
ENSGR0000214717.5 | zinc finger BED-type containing 1 |
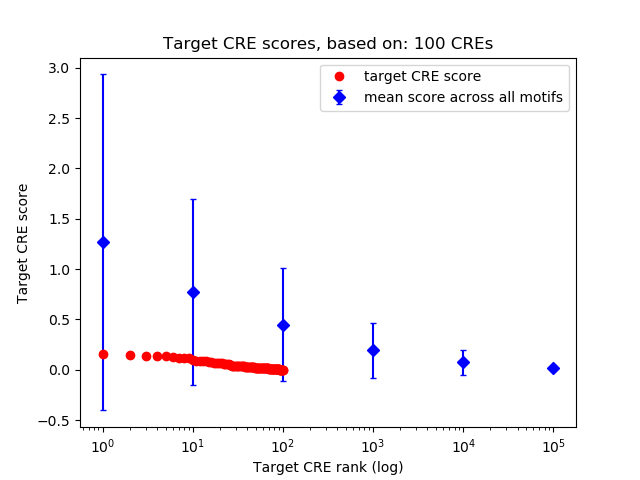
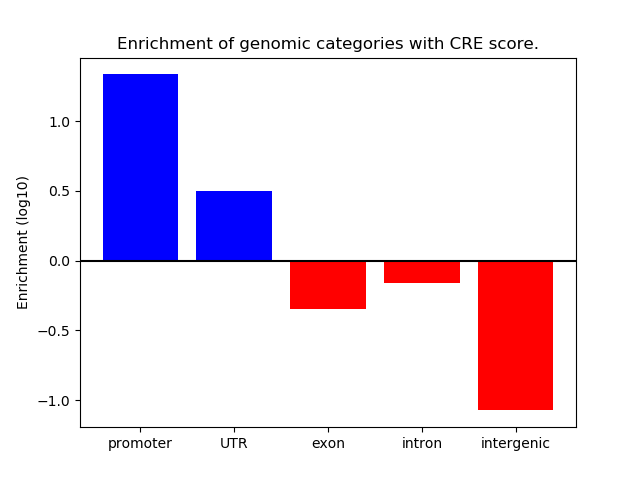
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_2402911_2403247 | ZBED1 | 15517 | 0.212992 | -0.80 | 9.0e-03 | Click! |
| chrX_2403256_2403492 | ZBED1 | 15222 | 0.213455 | -0.70 | 3.6e-02 | Click! |
| chrX_2418097_2418604 | ZBED1 | 246 | 0.605449 | 0.65 | 5.9e-02 | Click! |
| chrX_2402642_2402830 | ZBED1 | 15860 | 0.212450 | -0.64 | 6.5e-02 | Click! |
| chrX_2418812_2418977 | ZBED1 | 42 | 0.597697 | 0.46 | 2.1e-01 | Click! |
Activity of the ZBED1 motif across conditions
Conditions sorted by the z-value of the ZBED1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
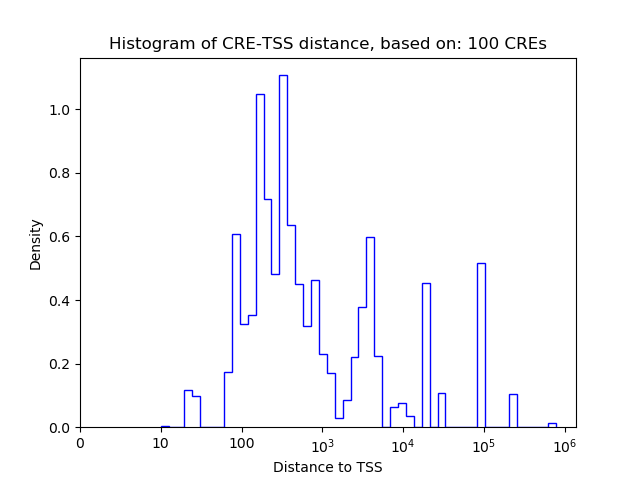
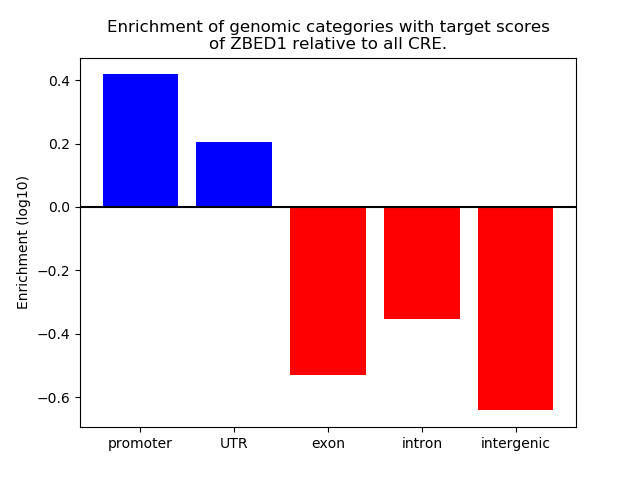
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_91862319_91862609 | 0.16 |
CCDC88C |
coiled-coil domain containing 88C |
21226 |
0.21 |
| chr1_36023479_36023699 | 0.15 |
NCDN |
neurochondrin |
180 |
0.68 |
| chr17_19093169_19093320 | 0.14 |
ENSG00000264940 |
. |
314 |
0.77 |
| chr15_90607371_90607522 | 0.14 |
ZNF710 |
zinc finger protein 710 |
3800 |
0.17 |
| chr2_677442_677606 | 0.13 |
TMEM18 |
transmembrane protein 18 |
85 |
0.8 |
| chr6_134273516_134273821 | 0.13 |
TBPL1 |
TBP-like 1 |
244 |
0.95 |
| chr17_19091594_19091745 | 0.12 |
ENSG00000263934 |
. |
340 |
0.74 |
| chr2_48010304_48010672 | 0.12 |
MSH6 |
mutS homolog 6 |
166 |
0.94 |
| chr5_140044777_140044956 | 0.12 |
WDR55 |
WD repeat domain 55 |
605 |
0.47 |
| chr10_31321494_31321708 | 0.10 |
ZNF438 |
zinc finger protein 438 |
735 |
0.75 |
| chr1_184723429_184723734 | 0.09 |
EDEM3 |
ER degradation enhancer, mannosidase alpha-like 3 |
120 |
0.97 |
| chr14_23235474_23235759 | 0.09 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
115 |
0.5 |
| chr18_74718505_74718656 | 0.08 |
MBP |
myelin basic protein |
3498 |
0.3 |
| chr1_6053732_6054005 | 0.08 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
1097 |
0.4 |
| chr21_35573901_35574398 | 0.08 |
ENSG00000266007 |
. |
103281 |
0.06 |
| chr20_18491437_18491588 | 0.08 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
2806 |
0.18 |
| chr4_152020832_152021050 | 0.07 |
RPS3A |
ribosomal protein S3A |
84 |
0.96 |
| chr17_30822708_30822859 | 0.07 |
RP11-466A19.1 |
|
229 |
0.88 |
| chr19_36630559_36630844 | 0.07 |
CAPNS1 |
calpain, small subunit 1 |
154 |
0.91 |
| chr4_100870352_100870894 | 0.06 |
RP11-15B17.1 |
|
540 |
0.59 |
| chr5_154237472_154237684 | 0.06 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
231 |
0.88 |
| chr19_2820167_2820484 | 0.06 |
ZNF554 |
zinc finger protein 554 |
377 |
0.76 |
| chr17_16290008_16290159 | 0.06 |
RP11-138I1.4 |
|
4100 |
0.15 |
| chr7_15501471_15501622 | 0.05 |
AGMO |
alkylglycerol monooxygenase |
95699 |
0.09 |
| chr17_18965457_18965608 | 0.05 |
ENSG00000265185 |
. |
307 |
0.8 |
| chr17_19015403_19015696 | 0.05 |
ENSG00000262202 |
. |
400 |
0.71 |
| chr12_112546663_112546943 | 0.04 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
23 |
0.97 |
| chr20_30467192_30467777 | 0.04 |
TTLL9 |
tubulin tyrosine ligase-like family, member 9 |
148 |
0.94 |
| chr20_3776560_3776785 | 0.04 |
CDC25B |
cell division cycle 25B |
190 |
0.91 |
| chr5_169308588_169308739 | 0.04 |
FAM196B |
family with sequence similarity 196, member B |
99081 |
0.07 |
| chr14_105267253_105268098 | 0.04 |
ZBTB42 |
zinc finger and BTB domain containing 42 |
425 |
0.78 |
| chr5_59284408_59284559 | 0.04 |
CTD-2254N19.1 |
|
32593 |
0.23 |
| chr2_182757184_182757414 | 0.04 |
SSFA2 |
sperm specific antigen 2 |
376 |
0.76 |
| chr1_30602704_30603087 | 0.04 |
ENSG00000222787 |
. |
245146 |
0.02 |
| chr5_70882830_70883043 | 0.04 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
188 |
0.96 |
| chr4_170541655_170542374 | 0.04 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
26 |
0.98 |
| chr19_3185557_3185759 | 0.03 |
NCLN |
nicalin |
60 |
0.96 |
| chr20_44002218_44002660 | 0.03 |
TP53TG5 |
TP53 target 5 |
4594 |
0.13 |
| chr12_114403396_114404100 | 0.03 |
RBM19 |
RNA binding motif protein 19 |
363 |
0.91 |
| chr19_40029876_40030364 | 0.03 |
EID2 |
EP300 interacting inhibitor of differentiation 2 |
750 |
0.48 |
| chr7_139025554_139026437 | 0.03 |
LUC7L2 |
LUC7-like 2 (S. cerevisiae) |
111 |
0.49 |
| chr5_149380291_149380778 | 0.03 |
TIGD6 |
tigger transposable element derived 6 |
196 |
0.56 |
| chr12_865415_865972 | 0.03 |
WNK1 |
WNK lysine deficient protein kinase 1 |
2961 |
0.28 |
| chr8_39807185_39807336 | 0.03 |
IDO2 |
indoleamine 2,3-dioxygenase 2 |
575 |
0.73 |
| chr14_52329027_52329412 | 0.02 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
1177 |
0.52 |
| chr7_5571391_5571891 | 0.02 |
ACTB |
actin, beta |
1301 |
0.32 |
| chr4_914174_914325 | 0.02 |
GAK |
cyclin G associated kinase |
4726 |
0.17 |
| chr10_42738908_42739069 | 0.02 |
ENSG00000264398 |
. |
8565 |
0.27 |
| chr16_68267852_68268003 | 0.02 |
ESRP2 |
epithelial splicing regulatory protein 2 |
2108 |
0.13 |
| chr4_77068973_77069571 | 0.02 |
NUP54 |
nucleoporin 54kDa |
278 |
0.9 |
| chr19_1268552_1268973 | 0.02 |
CIRBP |
cold inducible RNA binding protein |
568 |
0.5 |
| chr2_86849319_86850125 | 0.02 |
RNF103-CHMP3 |
RNF103-CHMP3 readthrough |
429 |
0.72 |
| chr14_23292100_23292804 | 0.02 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
168 |
0.82 |
| chr1_20126563_20126899 | 0.02 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
321 |
0.87 |
| chr1_152019828_152020543 | 0.02 |
S100A11 |
S100 calcium binding protein A11 |
10674 |
0.15 |
| chr8_26370640_26370791 | 0.02 |
PNMA2 |
paraneoplastic Ma antigen 2 |
893 |
0.46 |
| chr6_74232761_74232916 | 0.02 |
RP11-505P4.7 |
|
503 |
0.51 |
| chr5_5422874_5423110 | 0.02 |
KIAA0947 |
KIAA0947 |
185 |
0.97 |
| chr20_40246786_40247121 | 0.02 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
61 |
0.98 |
| chr3_47516346_47517336 | 0.02 |
SCAP |
SREBF chaperone |
246 |
0.9 |
| chr15_100105137_100105437 | 0.01 |
MEF2A |
myocyte enhancer factor 2A |
335 |
0.91 |
| chr4_54927912_54928194 | 0.01 |
AC110792.1 |
HCG2027126; Uncharacterized protein |
840 |
0.56 |
| chr12_57915744_57916361 | 0.01 |
MBD6 |
methyl-CpG binding domain protein 6 |
475 |
0.55 |
| chr19_44488599_44488754 | 0.01 |
ZNF155 |
zinc finger protein 155 |
303 |
0.82 |
| chr5_148929598_148930007 | 0.01 |
CSNK1A1 |
casein kinase 1, alpha 1 |
60 |
0.96 |
| chr6_136571677_136572089 | 0.01 |
MTFR2 |
mitochondrial fission regulator 2 |
421 |
0.85 |
| chr2_148603046_148603645 | 0.01 |
ACVR2A |
activin A receptor, type IIA |
1160 |
0.53 |
| chr4_44680371_44680949 | 0.01 |
YIPF7 |
Yip1 domain family, member 7 |
87 |
0.68 |
| chr3_10269933_10270194 | 0.01 |
ENSG00000222348 |
. |
11167 |
0.12 |
| chr19_45972718_45973011 | 0.01 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
270 |
0.85 |
| chr17_76136908_76137080 | 0.01 |
C17orf99 |
chromosome 17 open reading frame 99 |
5440 |
0.12 |
| chr11_17298561_17298964 | 0.01 |
NUCB2 |
nucleobindin 2 |
192 |
0.95 |
| chr18_23669563_23669744 | 0.01 |
SS18 |
synovial sarcoma translocation, chromosome 18 |
407 |
0.89 |
| chr8_56802055_56802342 | 0.01 |
RP11-318K15.2 |
|
3956 |
0.17 |
| chrX_152927870_152928027 | 0.01 |
ENSG00000244335 |
. |
3733 |
0.13 |
| chr14_92330770_92330921 | 0.01 |
TC2N |
tandem C2 domains, nuclear |
3028 |
0.27 |
| chr20_62669282_62669575 | 0.01 |
ZNF512B |
zinc finger protein 512B |
510 |
0.62 |
| chr1_25446753_25447069 | 0.01 |
RP4-706G24.1 |
|
87719 |
0.07 |
| chr3_195269115_195270107 | 0.01 |
PPP1R2 |
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
565 |
0.63 |
| chr22_28115426_28115577 | 0.01 |
RP11-375H17.1 |
|
3033 |
0.37 |
| chr2_28613525_28613857 | 0.01 |
FOSL2 |
FOS-like antigen 2 |
1978 |
0.28 |
| chr20_46414640_46415337 | 0.01 |
SULF2 |
sulfatase 2 |
310 |
0.92 |
| chr1_110881174_110881447 | 0.01 |
RBM15 |
RNA binding motif protein 15 |
182 |
0.77 |
| chr21_47745314_47745842 | 0.01 |
PCNT |
pericentrin |
1542 |
0.26 |
| chr12_122124988_122125315 | 0.01 |
MORN3 |
MORN repeat containing 3 |
17591 |
0.17 |
| chr10_22629025_22629540 | 0.01 |
SPAG6 |
sperm associated antigen 6 |
5117 |
0.17 |
| chr12_122516325_122516536 | 0.01 |
MLXIP |
MLX interacting protein |
198 |
0.95 |
| chr2_123651625_123651776 | 0.01 |
ENSG00000221689 |
. |
788091 |
0.0 |
| chr2_98708076_98708227 | 0.01 |
VWA3B |
von Willebrand factor A domain containing 3B |
4452 |
0.31 |
| chr19_8922272_8922597 | 0.00 |
CTD-2529P6.3 |
|
9743 |
0.14 |
| chr1_46908336_46908909 | 0.00 |
RP5-1109J22.1 |
|
10821 |
0.18 |
| chr6_124123668_124123827 | 0.00 |
NKAIN2 |
Na+/K+ transporting ATPase interacting 2 |
1539 |
0.46 |
| chr1_205561203_205561483 | 0.00 |
ENSG00000206762 |
. |
2826 |
0.21 |
| chr5_76382528_76383471 | 0.00 |
ZBED3 |
zinc finger, BED-type containing 3 |
9 |
0.67 |
| chr7_6272035_6272186 | 0.00 |
ENSG00000202273 |
. |
19739 |
0.16 |
| chr18_44526809_44527253 | 0.00 |
KATNAL2 |
katanin p60 subunit A-like 2 |
213 |
0.91 |
| chr3_129393451_129393602 | 0.00 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
14049 |
0.21 |
| chr19_9929067_9929467 | 0.00 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
238 |
0.8 |
| chr3_100280033_100280184 | 0.00 |
GPR128 |
G protein-coupled receptor 128 |
48325 |
0.14 |
| chr20_52789339_52789490 | 0.00 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
774 |
0.72 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032142 | four-way junction DNA binding(GO:0000400) single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |