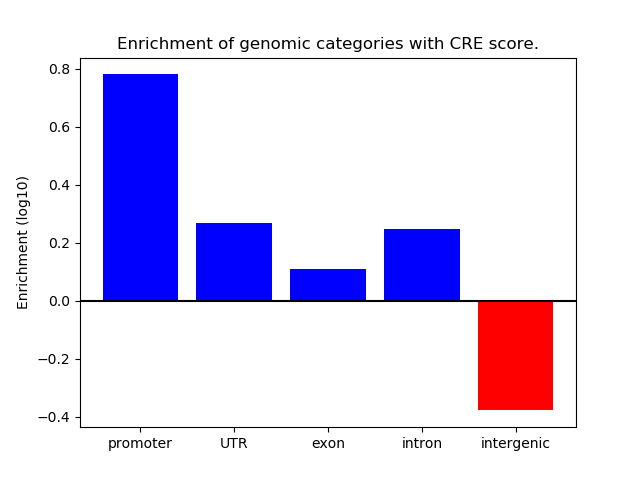
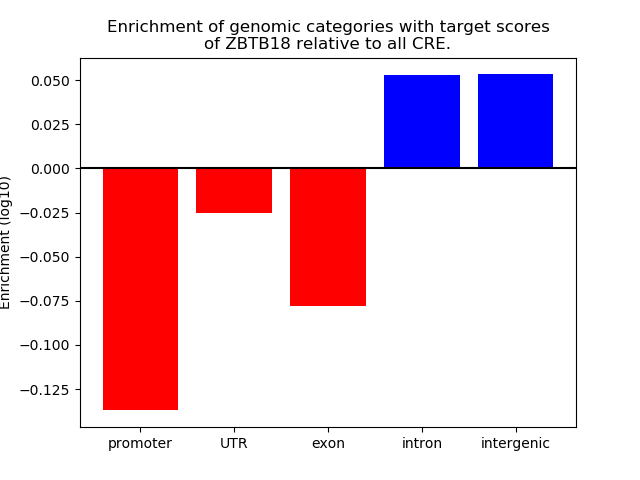
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
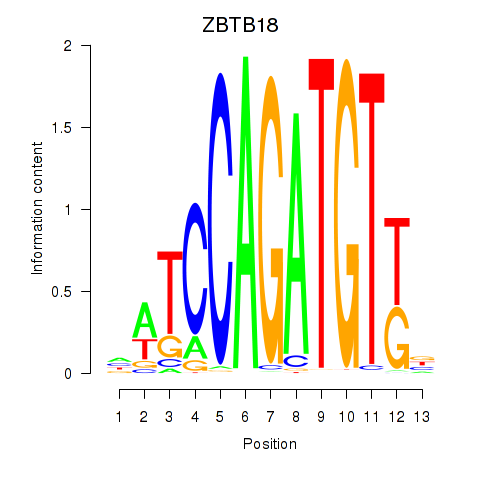
Results for ZBTB18
Z-value: 1.91
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
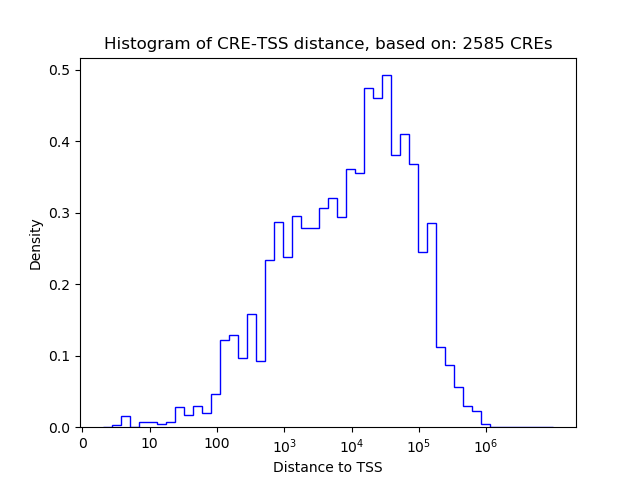
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_244122588_244122739 | ZBTB18 | 91922 | 0.077504 | 0.69 | 4.0e-02 | Click! |
| chr1_244210323_244211450 | ZBTB18 | 3699 | 0.266547 | -0.37 | 3.3e-01 | Click! |
| chr1_244217383_244217534 | ZBTB18 | 2873 | 0.289025 | -0.35 | 3.6e-01 | Click! |
| chr1_244214544_244214695 | ZBTB18 | 34 | 0.980846 | 0.34 | 3.7e-01 | Click! |
| chr1_244215459_244215610 | ZBTB18 | 949 | 0.610254 | -0.34 | 3.7e-01 | Click! |
Activity of the ZBTB18 motif across conditions
Conditions sorted by the z-value of the ZBTB18 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
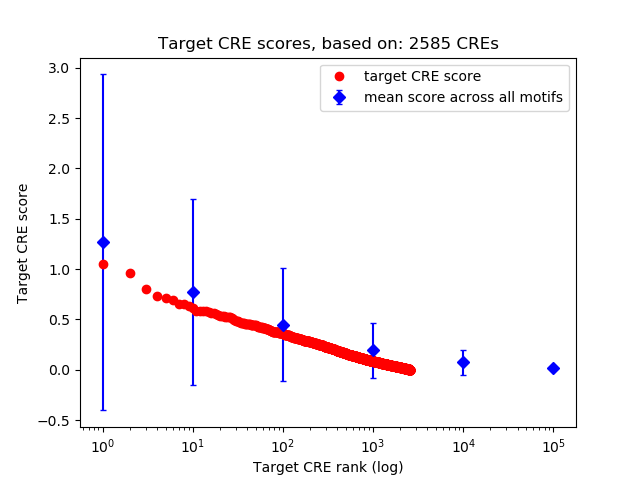
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_54379595_54379934 | 1.06 |
RP11-834C11.12 |
|
135 |
0.72 |
| chr2_218866425_218866801 | 0.97 |
TNS1 |
tensin 1 |
1105 |
0.51 |
| chrX_19688369_19688923 | 0.80 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
166 |
0.97 |
| chr1_232803371_232803522 | 0.74 |
ENSG00000238382 |
. |
32504 |
0.22 |
| chr21_46891372_46891858 | 0.71 |
COL18A1 |
collagen, type XVIII, alpha 1 |
16191 |
0.18 |
| chr4_138452558_138452766 | 0.69 |
PCDH18 |
protocadherin 18 |
903 |
0.76 |
| chr12_63194841_63194992 | 0.65 |
ENSG00000200296 |
. |
49765 |
0.16 |
| chr2_163099691_163099877 | 0.65 |
FAP |
fibroblast activation protein, alpha |
182 |
0.96 |
| chr1_16343782_16344178 | 0.63 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
340 |
0.8 |
| chr12_108983093_108983382 | 0.62 |
TMEM119 |
transmembrane protein 119 |
8547 |
0.14 |
| chr4_148402515_148403207 | 0.59 |
EDNRA |
endothelin receptor type A |
777 |
0.76 |
| chr17_71039690_71039871 | 0.58 |
SLC39A11 |
solute carrier family 39, member 11 |
46123 |
0.17 |
| chr3_190812364_190812515 | 0.58 |
OSTN |
osteocrin |
104591 |
0.07 |
| chr2_20291694_20292044 | 0.58 |
RP11-644K8.1 |
|
39974 |
0.13 |
| chr14_61110506_61110736 | 0.58 |
SIX1 |
SIX homeobox 1 |
5559 |
0.23 |
| chr7_116167584_116167883 | 0.57 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
1386 |
0.37 |
| chr8_17308069_17308220 | 0.57 |
MTMR7 |
myotubularin related protein 7 |
37308 |
0.17 |
| chr2_175202799_175202950 | 0.56 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
723 |
0.62 |
| chr8_40002568_40002719 | 0.55 |
C8orf4 |
chromosome 8 open reading frame 4 |
8346 |
0.3 |
| chr3_61580479_61580865 | 0.54 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
33087 |
0.24 |
| chr12_102821317_102821468 | 0.54 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
51037 |
0.17 |
| chr4_138626508_138626740 | 0.53 |
PCDH18 |
protocadherin 18 |
172976 |
0.04 |
| chr5_122615011_122615162 | 0.53 |
CEP120 |
centrosomal protein 120kDa |
143911 |
0.04 |
| chr17_64536538_64536785 | 0.53 |
ENSG00000252685 |
. |
97728 |
0.07 |
| chr7_30357859_30358186 | 0.53 |
ENSG00000207771 |
. |
28612 |
0.17 |
| chr4_135288096_135288247 | 0.52 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
165268 |
0.04 |
| chr15_60687226_60687377 | 0.52 |
ANXA2 |
annexin A2 |
2236 |
0.4 |
| chr19_41106605_41106828 | 0.51 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
561 |
0.68 |
| chr16_65154176_65154454 | 0.50 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
1518 |
0.59 |
| chr4_154506236_154506387 | 0.49 |
KIAA0922 |
KIAA0922 |
29263 |
0.2 |
| chr17_70662330_70662481 | 0.49 |
ENSG00000200783 |
. |
2114 |
0.41 |
| chr4_140808041_140808242 | 0.48 |
MAML3 |
mastermind-like 3 (Drosophila) |
3065 |
0.36 |
| chr11_7911988_7912218 | 0.47 |
ENSG00000252769 |
. |
1277 |
0.37 |
| chr6_2385878_2386080 | 0.47 |
ENSG00000266252 |
. |
23890 |
0.26 |
| chr12_13326265_13326502 | 0.47 |
EMP1 |
epithelial membrane protein 1 |
23267 |
0.21 |
| chr15_48935948_48936126 | 0.46 |
FBN1 |
fibrillin 1 |
1881 |
0.46 |
| chr21_29827177_29827328 | 0.46 |
ENSG00000251894 |
. |
288278 |
0.01 |
| chr5_148521385_148522083 | 0.46 |
ABLIM3 |
actin binding LIM protein family, member 3 |
124 |
0.96 |
| chr6_45500446_45500597 | 0.46 |
RUNX2 |
runt-related transcription factor 2 |
110299 |
0.06 |
| chr2_151333058_151333209 | 0.46 |
RND3 |
Rho family GTPase 3 |
8763 |
0.33 |
| chr13_36366182_36366333 | 0.46 |
DCLK1 |
doublecortin-like kinase 1 |
63522 |
0.15 |
| chr1_114522595_114522893 | 0.46 |
OLFML3 |
olfactomedin-like 3 |
49 |
0.97 |
| chr2_28211010_28211161 | 0.45 |
ENSG00000265321 |
. |
8149 |
0.25 |
| chr1_198873027_198873178 | 0.45 |
ENSG00000207759 |
. |
44820 |
0.17 |
| chr7_98734981_98735368 | 0.45 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
6468 |
0.25 |
| chr11_123065103_123065500 | 0.45 |
CLMP |
CXADR-like membrane protein |
688 |
0.71 |
| chr4_56731831_56731982 | 0.44 |
EXOC1 |
exocyst complex component 1 |
12001 |
0.19 |
| chr5_31955148_31955299 | 0.44 |
ENSG00000266243 |
. |
18958 |
0.21 |
| chr17_77019963_77021139 | 0.44 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
208 |
0.91 |
| chr1_235998951_235999123 | 0.44 |
LYST-AS1 |
LYST antisense RNA 1 |
3746 |
0.21 |
| chr7_120969496_120970092 | 0.44 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
737 |
0.77 |
| chr11_63767057_63767982 | 0.44 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
13205 |
0.11 |
| chr5_58655810_58656046 | 0.43 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
3127 |
0.41 |
| chr1_177980584_177980776 | 0.43 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
41332 |
0.2 |
| chr7_127307743_127307970 | 0.43 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
15622 |
0.17 |
| chr2_225577018_225577169 | 0.43 |
DOCK10 |
dedicator of cytokinesis 10 |
74742 |
0.12 |
| chr10_101548471_101548669 | 0.42 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
6050 |
0.22 |
| chr18_499765_500605 | 0.42 |
COLEC12 |
collectin sub-family member 12 |
537 |
0.79 |
| chr9_137536456_137536648 | 0.42 |
COL5A1 |
collagen, type V, alpha 1 |
2932 |
0.28 |
| chr2_160997260_160997411 | 0.42 |
ITGB6 |
integrin, beta 6 |
59428 |
0.14 |
| chr4_119248841_119248992 | 0.42 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
25242 |
0.23 |
| chr5_42683843_42683994 | 0.42 |
CCDC152 |
coiled-coil domain containing 152 |
72985 |
0.11 |
| chr18_74797223_74797374 | 0.41 |
MBP |
myelin basic protein |
19919 |
0.26 |
| chr7_98725447_98725838 | 0.41 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
16000 |
0.22 |
| chr7_27187795_27188438 | 0.41 |
HOXA6 |
homeobox A6 |
723 |
0.31 |
| chr9_82379166_82379317 | 0.41 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
57560 |
0.18 |
| chr22_29106631_29107007 | 0.41 |
CHEK2 |
checkpoint kinase 2 |
1100 |
0.47 |
| chr14_37050633_37050993 | 0.40 |
NKX2-8 |
NK2 homeobox 8 |
999 |
0.51 |
| chr1_109247037_109247211 | 0.40 |
FNDC7 |
fibronectin type III domain containing 7 |
8155 |
0.16 |
| chr13_98905877_98906028 | 0.40 |
RP11-573N10.1 |
|
20048 |
0.17 |
| chr1_17306478_17306650 | 0.40 |
MFAP2 |
microfibrillar-associated protein 2 |
609 |
0.57 |
| chr10_63402177_63402609 | 0.39 |
C10orf107 |
chromosome 10 open reading frame 107 |
20326 |
0.24 |
| chr1_219523892_219524043 | 0.38 |
RP11-135J2.4 |
|
176664 |
0.03 |
| chr2_192067901_192068052 | 0.38 |
MYO1B |
myosin IB |
41935 |
0.16 |
| chr1_87523622_87523774 | 0.38 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
64933 |
0.11 |
| chr15_96878792_96878943 | 0.38 |
ENSG00000222651 |
. |
2377 |
0.22 |
| chr8_119499637_119499788 | 0.38 |
SAMD12 |
sterile alpha motif domain containing 12 |
93409 |
0.1 |
| chr5_15914551_15914890 | 0.38 |
ENSG00000216077 |
. |
20571 |
0.27 |
| chr17_47654167_47654432 | 0.38 |
NXPH3 |
neurexophilin 3 |
815 |
0.39 |
| chr8_95254080_95254242 | 0.38 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
18837 |
0.21 |
| chr13_48093959_48094110 | 0.38 |
ENSG00000244521 |
. |
60665 |
0.16 |
| chr2_205125099_205125250 | 0.38 |
PARD3B |
par-3 family cell polarity regulator beta |
285342 |
0.01 |
| chr10_95241473_95241734 | 0.37 |
MYOF |
myoferlin |
348 |
0.88 |
| chr5_123991118_123991269 | 0.37 |
RP11-436H11.2 |
|
73331 |
0.09 |
| chr8_123101527_123101678 | 0.37 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
447926 |
0.01 |
| chr10_126416522_126416673 | 0.37 |
FAM53B |
family with sequence similarity 53, member B |
14942 |
0.16 |
| chr8_49468858_49469448 | 0.37 |
RP11-770E5.1 |
|
5026 |
0.35 |
| chr2_46076778_46077115 | 0.37 |
PRKCE |
protein kinase C, epsilon |
151095 |
0.04 |
| chr11_65084292_65084443 | 0.37 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
146 |
0.93 |
| chr3_123519098_123519249 | 0.37 |
MYLK |
myosin light chain kinase |
6485 |
0.22 |
| chr11_68038557_68038708 | 0.37 |
C11orf24 |
chromosome 11 open reading frame 24 |
783 |
0.67 |
| chr2_237551540_237552008 | 0.37 |
ACKR3 |
atypical chemokine receptor 3 |
73490 |
0.12 |
| chr12_70272871_70273022 | 0.37 |
MYRFL |
myelin regulatory factor-like |
47409 |
0.15 |
| chr12_3004539_3004690 | 0.37 |
TULP3 |
tubby like protein 3 |
4541 |
0.13 |
| chr2_98566923_98567218 | 0.36 |
ENSG00000238719 |
. |
37490 |
0.17 |
| chr21_40984409_40984913 | 0.36 |
C21orf88 |
chromosome 21 open reading frame 88 |
87 |
0.97 |
| chr10_30248594_30248745 | 0.36 |
KIAA1462 |
KIAA1462 |
99784 |
0.08 |
| chr4_81869449_81869600 | 0.36 |
BMP3 |
bone morphogenetic protein 3 |
82595 |
0.11 |
| chr1_14033707_14033883 | 0.36 |
PRDM2 |
PR domain containing 2, with ZNF domain |
2445 |
0.3 |
| chr14_69419003_69419387 | 0.36 |
ACTN1 |
actinin, alpha 1 |
4930 |
0.26 |
| chr10_33964074_33964254 | 0.36 |
NRP1 |
neuropilin 1 |
338974 |
0.01 |
| chr4_141229896_141230299 | 0.36 |
ENSG00000252300 |
. |
33831 |
0.16 |
| chr15_89251638_89251802 | 0.36 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
69536 |
0.09 |
| chr1_223314396_223314551 | 0.36 |
TLR5 |
toll-like receptor 5 |
2107 |
0.46 |
| chr6_101850561_101850712 | 0.35 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
3482 |
0.4 |
| chrX_48324188_48324448 | 0.35 |
SLC38A5 |
solute carrier family 38, member 5 |
1562 |
0.27 |
| chr15_76000821_76000972 | 0.35 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
4293 |
0.11 |
| chr7_81254313_81254464 | 0.35 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
137924 |
0.05 |
| chr15_96703759_96703910 | 0.35 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
62450 |
0.14 |
| chr3_151903763_151903914 | 0.35 |
MBNL1 |
muscleblind-like splicing regulator 1 |
81991 |
0.1 |
| chr6_136256925_136257180 | 0.35 |
ENSG00000238921 |
. |
21159 |
0.21 |
| chr12_57529324_57529537 | 0.34 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
3508 |
0.13 |
| chr8_128938263_128938598 | 0.34 |
TMEM75 |
transmembrane protein 75 |
22161 |
0.19 |
| chr7_78912183_78912334 | 0.34 |
ENSG00000212482 |
. |
60430 |
0.15 |
| chr7_47367053_47367431 | 0.34 |
TNS3 |
tensin 3 |
47326 |
0.2 |
| chr3_53106938_53107738 | 0.34 |
ENSG00000266635 |
. |
20324 |
0.16 |
| chr15_82185842_82186054 | 0.34 |
ENSG00000222521 |
. |
150687 |
0.04 |
| chr4_124981640_124982053 | 0.34 |
ANKRD50 |
ankyrin repeat domain 50 |
652041 |
0.0 |
| chr1_36104081_36104260 | 0.34 |
PSMB2 |
proteasome (prosome, macropain) subunit, beta type, 2 |
3275 |
0.22 |
| chr7_76145242_76145393 | 0.33 |
UPK3B |
uroplakin 3B |
5385 |
0.15 |
| chr7_42645149_42645379 | 0.33 |
C7orf25 |
chromosome 7 open reading frame 25 |
306245 |
0.01 |
| chr1_38022608_38022888 | 0.33 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
141 |
0.94 |
| chr16_64927185_64927365 | 0.33 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
166306 |
0.04 |
| chr6_16352044_16352195 | 0.33 |
ENSG00000265642 |
. |
76635 |
0.1 |
| chr9_123698083_123698281 | 0.33 |
TRAF1 |
TNF receptor-associated factor 1 |
6731 |
0.23 |
| chr2_97189877_97190028 | 0.33 |
RP11-363D14.1 |
|
8091 |
0.18 |
| chr16_87802522_87802699 | 0.32 |
KLHDC4 |
kelch domain containing 4 |
3028 |
0.24 |
| chr1_228134561_228135379 | 0.32 |
WNT9A |
wingless-type MMTV integration site family, member 9A |
629 |
0.71 |
| chr17_15160216_15160367 | 0.32 |
RP11-849N15.1 |
|
4041 |
0.17 |
| chr7_6298758_6298909 | 0.32 |
CYTH3 |
cytohesin 3 |
13442 |
0.18 |
| chr22_40490630_40490805 | 0.32 |
TNRC6B |
trinucleotide repeat containing 6B |
49239 |
0.13 |
| chr15_49715580_49715731 | 0.32 |
FGF7 |
fibroblast growth factor 7 |
198 |
0.96 |
| chr19_39919352_39919568 | 0.32 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
4049 |
0.09 |
| chr1_94793146_94793437 | 0.32 |
ARHGAP29 |
Rho GTPase activating protein 29 |
90102 |
0.08 |
| chr9_16828717_16828972 | 0.32 |
BNC2 |
basonuclin 2 |
3442 |
0.37 |
| chr19_10337313_10337464 | 0.32 |
ENSG00000264266 |
. |
3701 |
0.1 |
| chr9_131960482_131960899 | 0.32 |
RP11-247A12.2 |
|
2904 |
0.19 |
| chr10_49811984_49812175 | 0.32 |
ARHGAP22 |
Rho GTPase activating protein 22 |
918 |
0.63 |
| chr6_39760263_39760605 | 0.31 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
292 |
0.94 |
| chr16_83143220_83143371 | 0.31 |
CTD-3253I12.1 |
|
119392 |
0.06 |
| chr7_28076392_28076791 | 0.31 |
JAZF1 |
JAZF zinc finger 1 |
16173 |
0.26 |
| chr5_72114694_72114845 | 0.31 |
TNPO1 |
transportin 1 |
2270 |
0.27 |
| chr3_29507299_29507450 | 0.31 |
ENSG00000216169 |
. |
96462 |
0.08 |
| chr16_53120024_53120175 | 0.31 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
12973 |
0.23 |
| chr4_184338721_184338872 | 0.31 |
CDKN2AIP |
CDKN2A interacting protein |
26948 |
0.15 |
| chr2_150715873_150716024 | 0.31 |
ENSG00000207270 |
. |
248660 |
0.02 |
| chr10_68685803_68685995 | 0.31 |
LRRTM3 |
leucine rich repeat transmembrane neuronal 3 |
135 |
0.98 |
| chr2_190010666_190010817 | 0.31 |
ENSG00000264725 |
. |
12904 |
0.21 |
| chr10_18453497_18453677 | 0.31 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
23478 |
0.24 |
| chr8_59661345_59661496 | 0.31 |
ENSG00000201231 |
. |
65691 |
0.12 |
| chr20_49465299_49465721 | 0.31 |
ENSG00000265062 |
. |
11935 |
0.18 |
| chr4_142423380_142423531 | 0.31 |
IL15 |
interleukin 15 |
134297 |
0.05 |
| chr1_153517837_153518324 | 0.31 |
S100A4 |
S100 calcium binding protein A4 |
198 |
0.86 |
| chr1_34358710_34358861 | 0.31 |
RP5-1007G16.1 |
|
17718 |
0.22 |
| chr9_90812890_90813286 | 0.31 |
ENSG00000252299 |
. |
176096 |
0.03 |
| chr6_112652803_112652954 | 0.31 |
RFPL4B |
ret finger protein-like 4B |
15654 |
0.2 |
| chr8_123814017_123814168 | 0.30 |
RP11-44N11.1 |
|
5893 |
0.22 |
| chr2_201483563_201483714 | 0.30 |
AOX1 |
aldehyde oxidase 1 |
30741 |
0.18 |
| chr12_86198910_86199116 | 0.30 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
31335 |
0.22 |
| chr14_33296178_33296329 | 0.30 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
91196 |
0.09 |
| chr6_148608971_148609122 | 0.30 |
RP11-631F7.1 |
|
4502 |
0.23 |
| chr3_38216440_38216633 | 0.30 |
OXSR1 |
oxidative stress responsive 1 |
9540 |
0.16 |
| chr20_48183167_48183318 | 0.30 |
PTGIS |
prostaglandin I2 (prostacyclin) synthase |
1441 |
0.45 |
| chr8_107775219_107775370 | 0.30 |
ABRA |
actin-binding Rho activating protein |
7179 |
0.23 |
| chr8_38068343_38068494 | 0.30 |
BAG4 |
BCL2-associated athanogene 4 |
3307 |
0.15 |
| chr19_45751933_45752237 | 0.30 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
2431 |
0.2 |
| chr15_48845838_48845989 | 0.30 |
FBN1 |
fibrillin 1 |
92005 |
0.08 |
| chr4_126224474_126224625 | 0.30 |
FAT4 |
FAT atypical cadherin 4 |
13005 |
0.28 |
| chr12_122897921_122898072 | 0.29 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
9120 |
0.22 |
| chr1_246150409_246150560 | 0.29 |
RP11-83A16.1 |
|
46369 |
0.18 |
| chr4_71589225_71589655 | 0.29 |
RUFY3 |
RUN and FYVE domain containing 3 |
1068 |
0.4 |
| chr13_35962874_35963128 | 0.29 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
87831 |
0.09 |
| chr4_81116538_81116689 | 0.29 |
PRDM8 |
PR domain containing 8 |
2045 |
0.35 |
| chr19_11086584_11086796 | 0.29 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
8138 |
0.14 |
| chr11_62211430_62211631 | 0.29 |
CTD-2531D15.4 |
|
17334 |
0.11 |
| chr4_157867245_157867396 | 0.29 |
PDGFC |
platelet derived growth factor C |
24735 |
0.2 |
| chr1_172688052_172688203 | 0.29 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
59969 |
0.14 |
| chr4_103737426_103737626 | 0.29 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
9173 |
0.15 |
| chr6_152003026_152003177 | 0.29 |
ESR1 |
estrogen receptor 1 |
8530 |
0.26 |
| chr8_71070231_71070382 | 0.29 |
NCOA2 |
nuclear receptor coactivator 2 |
9611 |
0.26 |
| chr10_60235150_60235332 | 0.29 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
37659 |
0.2 |
| chr3_73835117_73835359 | 0.29 |
PDZRN3 |
PDZ domain containing ring finger 3 |
161147 |
0.04 |
| chr9_124461801_124462223 | 0.29 |
DAB2IP |
DAB2 interacting protein |
363 |
0.92 |
| chr3_78233290_78233441 | 0.29 |
ENSG00000222574 |
. |
22516 |
0.26 |
| chr7_26126830_26127131 | 0.28 |
ENSG00000266430 |
. |
27021 |
0.2 |
| chr7_98195950_98196101 | 0.28 |
NPTX2 |
neuronal pentraxin II |
50584 |
0.16 |
| chr3_11138169_11138337 | 0.28 |
HRH1 |
histamine receptor H1 |
40526 |
0.18 |
| chr19_1154555_1155129 | 0.28 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr9_79520083_79520617 | 0.28 |
PRUNE2 |
prune homolog 2 (Drosophila) |
651 |
0.82 |
| chr6_82755917_82756068 | 0.28 |
ENSG00000223044 |
. |
164165 |
0.04 |
| chr15_30112674_30113382 | 0.28 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr1_7130330_7130574 | 0.28 |
RP11-334N17.1 |
|
56113 |
0.15 |
| chr7_41227215_41227366 | 0.28 |
AC005160.3 |
|
412133 |
0.01 |
| chr3_156794251_156794402 | 0.28 |
ENSG00000222499 |
. |
59195 |
0.11 |
| chr3_11680037_11680346 | 0.28 |
VGLL4 |
vestigial like 4 (Drosophila) |
5207 |
0.24 |
| chr3_157157796_157157947 | 0.28 |
PTX3 |
pentraxin 3, long |
3293 |
0.33 |
| chr1_170645055_170645206 | 0.28 |
PRRX1 |
paired related homeobox 1 |
12052 |
0.28 |
| chr2_134324863_134325101 | 0.28 |
NCKAP5 |
NCK-associated protein 5 |
1049 |
0.62 |
| chr1_246024159_246024310 | 0.28 |
RP11-83A16.1 |
|
172619 |
0.03 |
| chr12_50410538_50410689 | 0.27 |
RACGAP1 |
Rac GTPase activating protein 1 |
110 |
0.95 |
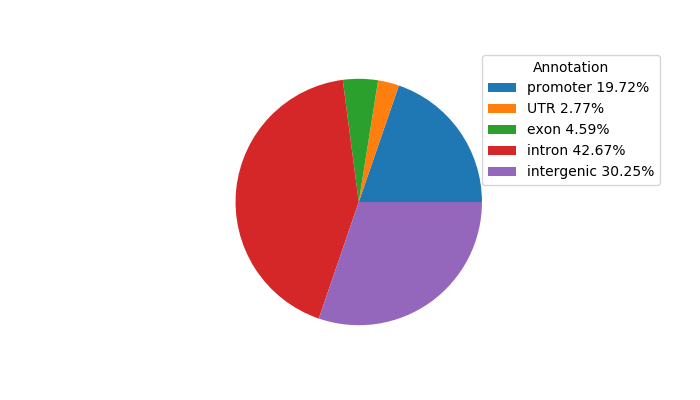
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.4 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.4 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.3 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.2 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.4 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.1 | GO:0031946 | regulation of glucocorticoid metabolic process(GO:0031943) regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.3 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0060840 | artery morphogenesis(GO:0048844) artery development(GO:0060840) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0052555 | positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.6 | GO:1901185 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |