Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
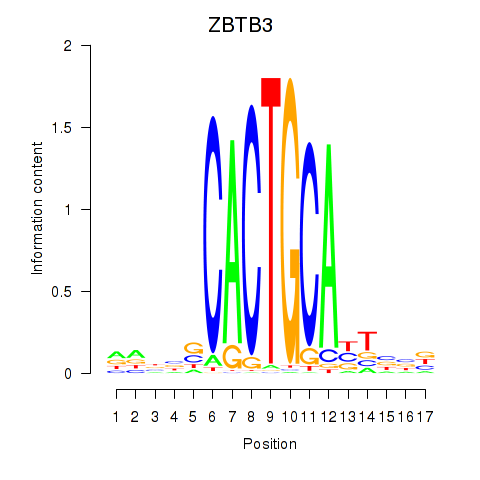
Results for ZBTB3
Z-value: 1.19
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.7 | zinc finger and BTB domain containing 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_62513178_62513329 | ZBTB3 | 8378 | 0.058854 | -0.56 | 1.2e-01 | Click! |
| chr11_62520878_62521029 | ZBTB3 | 678 | 0.379523 | -0.35 | 3.5e-01 | Click! |
| chr11_62521032_62521282 | ZBTB3 | 474 | 0.531828 | -0.33 | 3.8e-01 | Click! |
| chr11_62521434_62521626 | ZBTB3 | 101 | 0.891146 | -0.03 | 9.4e-01 | Click! |
| chr11_62521868_62522056 | ZBTB3 | 302 | 0.705283 | -0.00 | 9.9e-01 | Click! |
Activity of the ZBTB3 motif across conditions
Conditions sorted by the z-value of the ZBTB3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
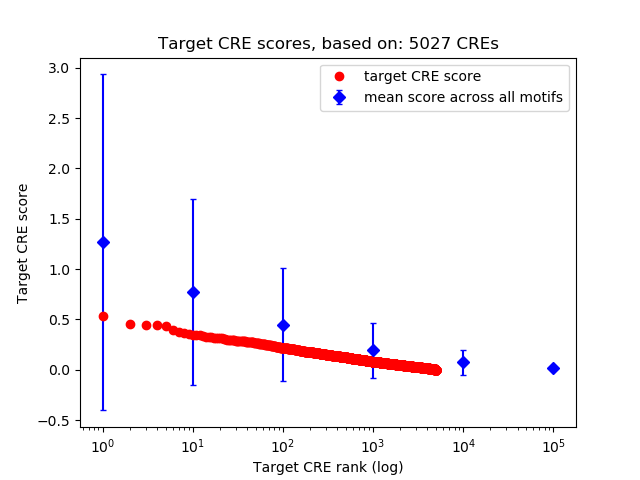
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_37882146_37882335 | 0.54 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
185 |
0.93 |
| chr13_31309966_31310295 | 0.45 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
485 |
0.88 |
| chrX_70317288_70317439 | 0.45 |
FOXO4 |
forkhead box O4 |
984 |
0.38 |
| chrX_153190817_153191535 | 0.45 |
ARHGAP4 |
Rho GTPase activating protein 4 |
522 |
0.6 |
| chr3_13057259_13057549 | 0.44 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
28868 |
0.23 |
| chr20_54987227_54988129 | 0.40 |
CASS4 |
Cas scaffolding protein family member 4 |
361 |
0.82 |
| chr17_73584096_73584435 | 0.37 |
MYO15B |
myosin XVB pseudogene |
2593 |
0.18 |
| chr12_122584467_122584714 | 0.37 |
MLXIP |
MLX interacting protein |
27837 |
0.17 |
| chr2_97529082_97529233 | 0.35 |
SEMA4C |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
5168 |
0.14 |
| chr1_50575757_50575939 | 0.35 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
470 |
0.86 |
| chr9_94668432_94668861 | 0.34 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
42516 |
0.2 |
| chrX_128914480_128914899 | 0.34 |
SASH3 |
SAM and SH3 domain containing 3 |
729 |
0.68 |
| chr6_24934206_24934437 | 0.33 |
FAM65B |
family with sequence similarity 65, member B |
1867 |
0.41 |
| chr6_136785760_136785911 | 0.33 |
MAP7 |
microtubule-associated protein 7 |
2178 |
0.39 |
| chrX_1653652_1653877 | 0.32 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
2236 |
0.34 |
| chr19_4541586_4541737 | 0.32 |
LRG1 |
leucine-rich alpha-2-glycoprotein 1 |
1175 |
0.29 |
| chr5_87797997_87798358 | 0.32 |
TMEM161B-AS1 |
TMEM161B antisense RNA 1 |
75665 |
0.11 |
| chrY_1603652_1603871 | 0.32 |
NA |
NA |
> 106 |
NA |
| chrX_13018542_13018927 | 0.32 |
TMSB4X |
thymosin beta 4, X-linked |
24957 |
0.19 |
| chr1_21916964_21917115 | 0.31 |
ENSG00000221781 |
. |
2002 |
0.27 |
| chr1_244488212_244488377 | 0.31 |
C1orf100 |
chromosome 1 open reading frame 100 |
27643 |
0.22 |
| chr11_93275392_93276314 | 0.30 |
SMCO4 |
single-pass membrane protein with coiled-coil domains 4 |
668 |
0.7 |
| chr5_175086058_175086209 | 0.30 |
HRH2 |
histamine receptor H2 |
1100 |
0.55 |
| chr17_27467519_27468040 | 0.30 |
RP11-321A17.4 |
|
278 |
0.55 |
| chr14_91696084_91696235 | 0.30 |
CTD-2547L24.3 |
HCG1816139; Uncharacterized protein |
12944 |
0.16 |
| chr19_54804071_54804222 | 0.30 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
27 |
0.95 |
| chr1_214484137_214484288 | 0.29 |
SMYD2 |
SET and MYND domain containing 2 |
20108 |
0.24 |
| chr1_40860301_40860452 | 0.29 |
SMAP2 |
small ArfGAP2 |
2131 |
0.29 |
| chrX_118827540_118827798 | 0.29 |
SEPT6 |
septin 6 |
336 |
0.88 |
| chr7_158496672_158497408 | 0.29 |
NCAPG2 |
non-SMC condensin II complex, subunit G2 |
417 |
0.87 |
| chr14_95652052_95652434 | 0.29 |
CTD-2240H23.2 |
|
52 |
0.97 |
| chr14_93119065_93119528 | 0.29 |
RIN3 |
Ras and Rab interactor 3 |
450 |
0.88 |
| chr17_62153670_62153821 | 0.29 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
53740 |
0.09 |
| chr6_139947376_139947865 | 0.29 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
251863 |
0.02 |
| chr7_71523639_71523917 | 0.29 |
ENSG00000201014 |
. |
144937 |
0.05 |
| chr1_40840894_40841045 | 0.29 |
SMAP2 |
small ArfGAP2 |
649 |
0.7 |
| chr7_127225793_127226035 | 0.28 |
GCC1 |
GRIP and coiled-coil domain containing 1 |
253 |
0.89 |
| chr8_145086499_145086675 | 0.28 |
SPATC1 |
spermatogenesis and centriole associated 1 |
5 |
0.83 |
| chr1_235530756_235531012 | 0.28 |
TBCE |
tubulin folding cofactor E |
139 |
0.96 |
| chr2_121102314_121103173 | 0.28 |
INHBB |
inhibin, beta B |
976 |
0.66 |
| chr7_37488572_37488804 | 0.28 |
ELMO1 |
engulfment and cell motility 1 |
101 |
0.97 |
| chr4_74965175_74965371 | 0.28 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
263 |
0.9 |
| chr14_21250158_21250382 | 0.28 |
RNASE6 |
ribonuclease, RNase A family, k6 |
1060 |
0.33 |
| chr3_187718091_187718280 | 0.28 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
152887 |
0.04 |
| chr16_29638502_29638653 | 0.27 |
ENSG00000266758 |
. |
27991 |
0.11 |
| chr5_141293636_141294105 | 0.27 |
KIAA0141 |
KIAA0141 |
9503 |
0.15 |
| chr3_46185157_46185308 | 0.27 |
CCR3 |
chemokine (C-C motif) receptor 3 |
19864 |
0.21 |
| chr6_134547435_134547586 | 0.27 |
ENSG00000238631 |
. |
19858 |
0.17 |
| chr3_129323468_129324020 | 0.27 |
PLXND1 |
plexin D1 |
1917 |
0.3 |
| chrX_129306905_129307398 | 0.27 |
RAB33A |
RAB33A, member RAS oncogene family |
1528 |
0.4 |
| chr3_125486908_125487059 | 0.27 |
ENSG00000221737 |
. |
22412 |
0.17 |
| chr7_38272697_38272848 | 0.26 |
STARD3NL |
STARD3 N-terminal like |
54775 |
0.17 |
| chr14_105527834_105528220 | 0.26 |
GPR132 |
G protein-coupled receptor 132 |
3740 |
0.23 |
| chr13_107572015_107572438 | 0.26 |
ENSG00000239050 |
. |
55148 |
0.17 |
| chr5_77806248_77806529 | 0.26 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
38586 |
0.21 |
| chr19_704286_704437 | 0.26 |
PALM |
paralemmin |
4592 |
0.12 |
| chr3_72226555_72227176 | 0.26 |
ENSG00000212070 |
. |
84714 |
0.11 |
| chr2_46497095_46497294 | 0.26 |
EPAS1 |
endothelial PAS domain protein 1 |
26553 |
0.23 |
| chr6_109805084_109805246 | 0.26 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
725 |
0.53 |
| chr13_99958033_99958184 | 0.26 |
GPR183 |
G protein-coupled receptor 183 |
1551 |
0.41 |
| chr11_121567136_121567530 | 0.26 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
106205 |
0.08 |
| chrX_48542477_48542733 | 0.25 |
WAS |
Wiskott-Aldrich syndrome |
437 |
0.75 |
| chr20_48322013_48322164 | 0.25 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
8327 |
0.2 |
| chr8_61851632_61851783 | 0.25 |
CLVS1 |
clavesin 1 |
118010 |
0.06 |
| chr11_29141848_29142035 | 0.25 |
ENSG00000211499 |
. |
156295 |
0.04 |
| chr5_114878889_114879040 | 0.25 |
FEM1C |
fem-1 homolog c (C. elegans) |
1627 |
0.42 |
| chr4_159092030_159092251 | 0.25 |
RP11-597D13.9 |
|
236 |
0.78 |
| chr15_33356813_33357043 | 0.25 |
FMN1 |
formin 1 |
3157 |
0.36 |
| chr3_43066068_43066219 | 0.25 |
FAM198A |
family with sequence similarity 198, member A |
45103 |
0.13 |
| chrX_65233867_65234034 | 0.25 |
ENSG00000207939 |
. |
4762 |
0.23 |
| chr5_102202140_102202291 | 0.25 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
395 |
0.92 |
| chr9_139260720_139260871 | 0.25 |
DNLZ |
DNL-type zinc finger |
2554 |
0.16 |
| chr4_186631734_186631914 | 0.24 |
ENSG00000207497 |
. |
703 |
0.7 |
| chr3_149194205_149194356 | 0.24 |
TM4SF4 |
transmembrane 4 L six family member 4 |
1794 |
0.39 |
| chr20_30640759_30640910 | 0.24 |
HCK |
hemopoietic cell kinase |
770 |
0.5 |
| chr9_80637867_80638081 | 0.24 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
7546 |
0.32 |
| chr3_50644095_50644246 | 0.24 |
CISH |
cytokine inducible SH2-containing protein |
5033 |
0.16 |
| chr1_151566174_151566325 | 0.23 |
SNX27 |
sorting nexin family member 27 |
18309 |
0.1 |
| chr1_235097453_235097604 | 0.23 |
ENSG00000239690 |
. |
57595 |
0.14 |
| chr11_64009123_64009509 | 0.23 |
FKBP2 |
FK506 binding protein 2, 13kDa |
96 |
0.89 |
| chr15_99500155_99500306 | 0.23 |
IGF1R |
insulin-like growth factor 1 receptor |
32401 |
0.16 |
| chr2_172877202_172877353 | 0.23 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
12787 |
0.23 |
| chr7_149572287_149572438 | 0.23 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
1230 |
0.35 |
| chr15_91446638_91446871 | 0.23 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
220 |
0.86 |
| chr2_99388395_99388567 | 0.23 |
ENSG00000201070 |
. |
10392 |
0.22 |
| chr1_39681126_39681366 | 0.23 |
RP11-416A14.1 |
|
9062 |
0.18 |
| chr1_209930383_209930698 | 0.23 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
1016 |
0.45 |
| chr6_26157499_26157809 | 0.23 |
HIST1H2BD |
histone cluster 1, H2bd |
695 |
0.35 |
| chr20_10455686_10455837 | 0.22 |
SLX4IP |
SLX4 interacting protein |
39810 |
0.16 |
| chr10_22896358_22896509 | 0.22 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
15791 |
0.28 |
| chr1_235114255_235114474 | 0.22 |
ENSG00000239690 |
. |
74431 |
0.11 |
| chr8_52810422_52811259 | 0.22 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
756 |
0.46 |
| chr1_244999063_244999557 | 0.22 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
392 |
0.85 |
| chr1_62208072_62208403 | 0.22 |
INADL |
InaD-like (Drosophila) |
88 |
0.98 |
| chr7_38352649_38352985 | 0.22 |
STARD3NL |
STARD3 N-terminal like |
134820 |
0.05 |
| chr16_85887051_85887202 | 0.22 |
IRF8 |
interferon regulatory factor 8 |
45283 |
0.1 |
| chr2_219235805_219236126 | 0.22 |
ENSG00000225062 |
. |
3407 |
0.11 |
| chr5_168217801_168217952 | 0.22 |
ENSG00000202345 |
. |
627 |
0.77 |
| chrX_13271711_13271862 | 0.22 |
GS1-600G8.3 |
|
56985 |
0.14 |
| chr14_56381006_56381157 | 0.22 |
ENSG00000212522 |
. |
154080 |
0.04 |
| chrX_70472547_70472698 | 0.22 |
ZMYM3 |
zinc finger, MYM-type 3 |
1357 |
0.36 |
| chr3_185870255_185870406 | 0.22 |
ETV5 |
ets variant 5 |
42223 |
0.17 |
| chr12_68772939_68773090 | 0.22 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
46853 |
0.17 |
| chr16_23159783_23160184 | 0.22 |
USP31 |
ubiquitin specific peptidase 31 |
608 |
0.78 |
| chr2_106754814_106755143 | 0.22 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
264 |
0.94 |
| chr1_89664351_89664632 | 0.22 |
GBP4 |
guanylate binding protein 4 |
124 |
0.96 |
| chr17_79197162_79197682 | 0.22 |
AZI1 |
5-azacytidine induced 1 |
623 |
0.55 |
| chr12_96502184_96502412 | 0.22 |
ENSG00000266889 |
. |
6271 |
0.22 |
| chr5_75471567_75471805 | 0.22 |
ENSG00000252833 |
. |
84060 |
0.09 |
| chr1_192777377_192777993 | 0.21 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
486 |
0.87 |
| chr1_161038546_161039545 | 0.21 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr6_163826222_163826373 | 0.21 |
QKI |
QKI, KH domain containing, RNA binding |
9378 |
0.32 |
| chrX_153978145_153979054 | 0.21 |
GAB3 |
GRB2-associated binding protein 3 |
733 |
0.55 |
| chr11_128161363_128161514 | 0.21 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
213851 |
0.02 |
| chr15_75081452_75081829 | 0.21 |
ENSG00000264386 |
. |
542 |
0.49 |
| chr16_85089458_85089881 | 0.21 |
KIAA0513 |
KIAA0513 |
7149 |
0.23 |
| chr1_111417492_111418003 | 0.21 |
CD53 |
CD53 molecule |
1971 |
0.34 |
| chr9_139267510_139268079 | 0.21 |
CARD9 |
caspase recruitment domain family, member 9 |
339 |
0.79 |
| chr1_100866510_100866661 | 0.21 |
ENSG00000216067 |
. |
22254 |
0.18 |
| chr7_75583164_75583315 | 0.21 |
POR |
P450 (cytochrome) oxidoreductase |
155 |
0.95 |
| chr12_53719733_53719917 | 0.21 |
AAAS |
achalasia, adrenocortical insufficiency, alacrimia |
1283 |
0.29 |
| chr17_9937060_9937211 | 0.21 |
GAS7 |
growth arrest-specific 7 |
2719 |
0.32 |
| chr9_100668829_100668981 | 0.21 |
C9orf156 |
chromosome 9 open reading frame 156 |
5848 |
0.17 |
| chr2_69271007_69271158 | 0.21 |
ANTXR1 |
anthrax toxin receptor 1 |
30553 |
0.17 |
| chr7_5862773_5862988 | 0.21 |
ZNF815P |
zinc finger protein 815, pseudogene |
42 |
0.97 |
| chr16_68122944_68123162 | 0.21 |
ENSG00000201850 |
. |
393 |
0.75 |
| chr11_67042666_67042837 | 0.21 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
8799 |
0.11 |
| chr12_53023939_53024090 | 0.21 |
KRT73 |
keratin 73 |
11671 |
0.11 |
| chr16_2932458_2932721 | 0.21 |
FLYWCH2 |
FLYWCH family member 2 |
598 |
0.55 |
| chr10_108596529_108596824 | 0.20 |
ENSG00000200626 |
. |
133103 |
0.06 |
| chr19_55765720_55766271 | 0.20 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
1142 |
0.27 |
| chr17_33863414_33863594 | 0.20 |
SLFN12L |
schlafen family member 12-like |
1376 |
0.29 |
| chr14_105553392_105553992 | 0.20 |
GPR132 |
G protein-coupled receptor 132 |
21910 |
0.17 |
| chr18_25755743_25755974 | 0.20 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
1552 |
0.58 |
| chr5_32709132_32709283 | 0.20 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
306 |
0.93 |
| chr5_176543501_176543652 | 0.20 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
16504 |
0.17 |
| chr22_46682638_46682789 | 0.20 |
GTSE1 |
G-2 and S-phase expressed 1 |
9925 |
0.15 |
| chr2_25517043_25517291 | 0.20 |
ENSG00000221445 |
. |
34423 |
0.16 |
| chr8_58779427_58779708 | 0.20 |
FAM110B |
family with sequence similarity 110, member B |
127546 |
0.06 |
| chr12_54746284_54746435 | 0.20 |
RP11-753H16.3 |
|
1086 |
0.24 |
| chr19_41311125_41311276 | 0.20 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
1847 |
0.24 |
| chrX_13108137_13108288 | 0.20 |
FAM9C |
family with sequence similarity 9, member C |
45411 |
0.18 |
| chr1_96985633_96985784 | 0.20 |
ENSG00000241992 |
. |
63057 |
0.15 |
| chr8_61880060_61880353 | 0.20 |
CLVS1 |
clavesin 1 |
89511 |
0.1 |
| chr7_28109764_28109915 | 0.19 |
JAZF1 |
JAZF zinc finger 1 |
1464 |
0.51 |
| chr3_37189437_37189588 | 0.19 |
ENSG00000199594 |
. |
7694 |
0.16 |
| chr1_26954743_26954935 | 0.19 |
ENSG00000238316 |
. |
14041 |
0.13 |
| chr3_142683549_142683700 | 0.19 |
RP11-372E1.6 |
|
285 |
0.46 |
| chr13_80809877_80810028 | 0.19 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
103842 |
0.08 |
| chr22_50523028_50523179 | 0.19 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
585 |
0.67 |
| chr4_8324999_8325150 | 0.19 |
HTRA3 |
HtrA serine peptidase 3 |
53570 |
0.12 |
| chr12_11926562_11926926 | 0.19 |
ETV6 |
ets variant 6 |
21309 |
0.26 |
| chr5_126187507_126187715 | 0.19 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
66309 |
0.11 |
| chr8_95956511_95956662 | 0.19 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
3385 |
0.17 |
| chr12_11807352_11807522 | 0.19 |
ETV6 |
ets variant 6 |
4649 |
0.31 |
| chr8_67575803_67576925 | 0.19 |
VCPIP1 |
valosin containing protein (p97)/p47 complex interacting protein 1 |
3088 |
0.17 |
| chr1_169763319_169763807 | 0.19 |
METTL18 |
methyltransferase like 18 |
267 |
0.75 |
| chr16_66864896_66865106 | 0.19 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
101 |
0.95 |
| chr6_159482531_159482682 | 0.19 |
TAGAP |
T-cell activation RhoGTPase activating protein |
16422 |
0.19 |
| chr5_173202634_173202785 | 0.19 |
ENSG00000263401 |
. |
45628 |
0.18 |
| chr12_29301342_29301647 | 0.19 |
FAR2 |
fatty acyl CoA reductase 2 |
542 |
0.83 |
| chr21_38283093_38283244 | 0.19 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
55605 |
0.1 |
| chr15_42867116_42867674 | 0.19 |
STARD9 |
StAR-related lipid transfer (START) domain containing 9 |
462 |
0.74 |
| chr8_23368073_23368224 | 0.19 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
18170 |
0.16 |
| chr14_70394671_70394822 | 0.19 |
SMOC1 |
SPARC related modular calcium binding 1 |
48603 |
0.16 |
| chr17_59487763_59488024 | 0.19 |
RP11-332H18.4 |
|
1023 |
0.34 |
| chr8_2151450_2151601 | 0.19 |
MYOM2 |
myomesin 2 |
158341 |
0.04 |
| chr15_58433772_58433923 | 0.19 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
2823 |
0.28 |
| chr1_222594201_222594352 | 0.18 |
ENSG00000222399 |
. |
82801 |
0.09 |
| chr10_29969861_29970012 | 0.18 |
ENSG00000222092 |
. |
30908 |
0.18 |
| chr6_155054889_155055250 | 0.18 |
SCAF8 |
SR-related CTD-associated factor 8 |
519 |
0.85 |
| chr6_136173249_136173683 | 0.18 |
PDE7B |
phosphodiesterase 7B |
632 |
0.81 |
| chr8_26298579_26298982 | 0.18 |
RP11-14I17.3 |
|
773 |
0.66 |
| chr13_41931400_41931551 | 0.18 |
ENSG00000223280 |
. |
2680 |
0.28 |
| chr11_18548581_18549104 | 0.18 |
TSG101 |
tumor susceptibility 101 |
63 |
0.97 |
| chr15_63340381_63340577 | 0.18 |
TPM1 |
tropomyosin 1 (alpha) |
86 |
0.94 |
| chr1_109395002_109395242 | 0.18 |
AKNAD1 |
AKNA domain containing 1 |
217 |
0.92 |
| chr10_81007808_81008068 | 0.18 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
58037 |
0.14 |
| chr17_38476195_38476719 | 0.18 |
RARA |
retinoic acid receptor, alpha |
1920 |
0.22 |
| chr16_28504630_28504781 | 0.18 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
611 |
0.54 |
| chr15_40699677_40699828 | 0.18 |
IVD |
isovaleryl-CoA dehydrogenase |
393 |
0.72 |
| chr13_24777681_24777832 | 0.18 |
ENSG00000252695 |
. |
41201 |
0.13 |
| chr1_207094900_207095227 | 0.18 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
149 |
0.94 |
| chr1_28503293_28503444 | 0.18 |
PTAFR |
platelet-activating factor receptor |
331 |
0.81 |
| chr4_170540985_170541480 | 0.18 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
440 |
0.86 |
| chr19_55768482_55768951 | 0.18 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
1579 |
0.19 |
| chr19_56132551_56132702 | 0.18 |
ZNF784 |
zinc finger protein 784 |
3306 |
0.09 |
| chr8_101225603_101226122 | 0.18 |
ENSG00000199667 |
. |
28496 |
0.14 |
| chr20_62219212_62219363 | 0.18 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
15479 |
0.09 |
| chr3_46248309_46248460 | 0.18 |
CCR1 |
chemokine (C-C motif) receptor 1 |
1503 |
0.46 |
| chr11_67138355_67138676 | 0.18 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
2693 |
0.12 |
| chr14_59104062_59104397 | 0.18 |
DACT1 |
dishevelled-binding antagonist of beta-catenin 1 |
545 |
0.85 |
| chr12_123200741_123200892 | 0.18 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
623 |
0.65 |
| chr2_98630005_98630391 | 0.18 |
TMEM131 |
transmembrane protein 131 |
17810 |
0.24 |
| chr1_110025821_110026323 | 0.18 |
ATXN7L2 |
ataxin 7-like 2 |
489 |
0.68 |
| chr12_31782840_31782991 | 0.18 |
METTL20 |
methyltransferase like 20 |
17179 |
0.15 |
| chr5_138940324_138940673 | 0.18 |
UBE2D2 |
ubiquitin-conjugating enzyme E2D 2 |
253 |
0.91 |
| chr6_42989627_42989814 | 0.18 |
RRP36 |
ribosomal RNA processing 36 homolog (S. cerevisiae) |
337 |
0.73 |
| chr17_42846307_42846458 | 0.18 |
ADAM11 |
ADAM metallopeptidase domain 11 |
9689 |
0.14 |
| chr1_159823893_159825316 | 0.18 |
C1orf204 |
chromosome 1 open reading frame 204 |
533 |
0.6 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.3 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.1 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0002830 | positive regulation of type 2 immune response(GO:0002830) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0060294 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.2 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.3 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0060431 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.2 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.0 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.0 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0035586 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) purinergic receptor activity(GO:0035586) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |