Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
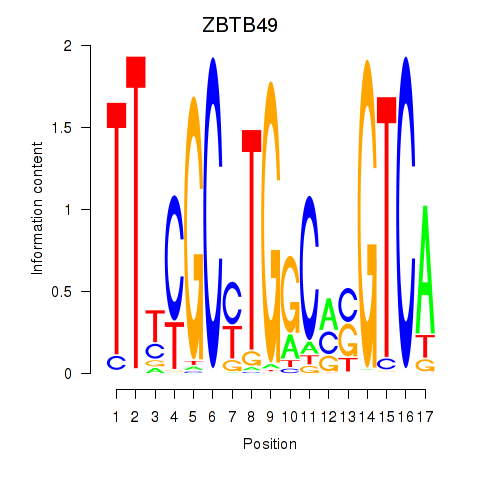
Results for ZBTB49
Z-value: 0.80
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | zinc finger and BTB domain containing 49 |
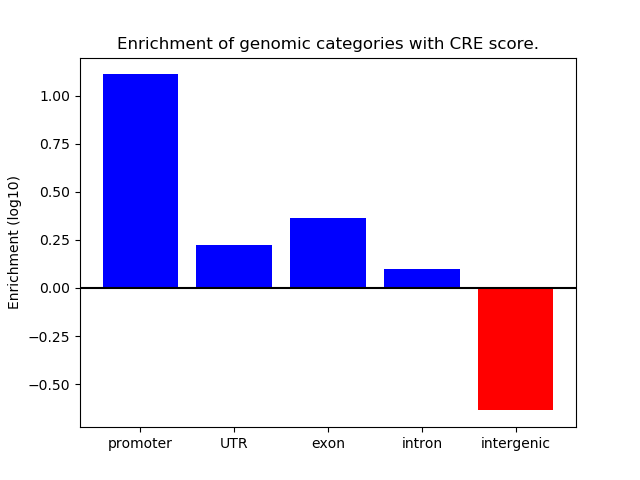
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_4316995_4317146 | ZBTB49 | 12716 | 0.176457 | 0.50 | 1.7e-01 | Click! |
| chr4_4316659_4316810 | ZBTB49 | 12380 | 0.177030 | 0.34 | 3.7e-01 | Click! |
| chr4_4292047_4292578 | ZBTB49 | 316 | 0.576045 | -0.10 | 7.9e-01 | Click! |
| chr4_4317457_4317608 | ZBTB49 | 13178 | 0.175678 | 0.03 | 9.3e-01 | Click! |
Activity of the ZBTB49 motif across conditions
Conditions sorted by the z-value of the ZBTB49 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
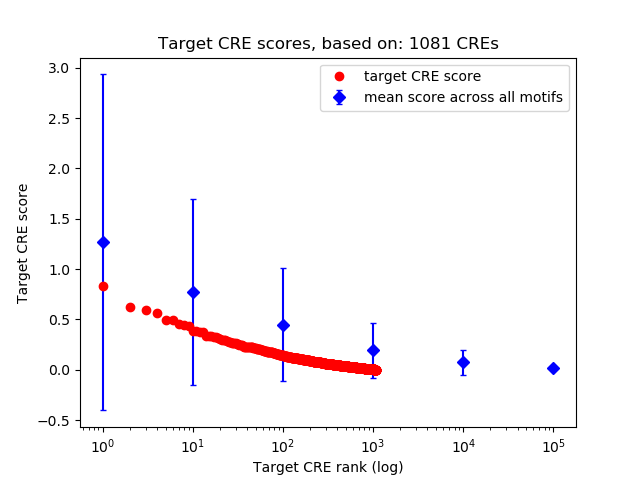
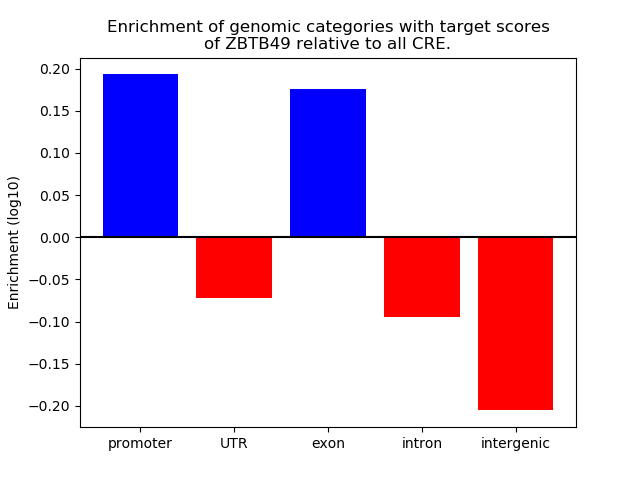
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_6481304_6482171 | 0.83 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr19_2084034_2084229 | 0.62 |
MOB3A |
MOB kinase activator 3A |
1260 |
0.31 |
| chr21_46340531_46340791 | 0.60 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
109 |
0.73 |
| chr12_15111636_15111984 | 0.56 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
2390 |
0.25 |
| chr17_3818740_3818909 | 0.50 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
970 |
0.51 |
| chr17_47819247_47819793 | 0.50 |
FAM117A |
family with sequence similarity 117, member A |
17631 |
0.14 |
| chr7_155150623_155150868 | 0.46 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
9884 |
0.18 |
| chr4_40207688_40207871 | 0.44 |
RHOH |
ras homolog family member H |
5815 |
0.23 |
| chr10_101290718_101290924 | 0.44 |
RP11-129J12.2 |
|
113 |
0.96 |
| chr19_42052588_42052799 | 0.39 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
3193 |
0.22 |
| chr19_35939406_35939567 | 0.38 |
FFAR2 |
free fatty acid receptor 2 |
283 |
0.85 |
| chr6_159465648_159465970 | 0.38 |
TAGAP |
T-cell activation RhoGTPase activating protein |
241 |
0.93 |
| chr3_53206269_53206420 | 0.37 |
PRKCD |
protein kinase C, delta |
7199 |
0.18 |
| chr8_8377701_8378015 | 0.34 |
ENSG00000263616 |
. |
62307 |
0.13 |
| chr19_18507519_18507670 | 0.34 |
LRRC25 |
leucine rich repeat containing 25 |
827 |
0.4 |
| chr8_27183037_27183778 | 0.33 |
PTK2B |
protein tyrosine kinase 2 beta |
326 |
0.9 |
| chr2_175459196_175459684 | 0.33 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
3053 |
0.24 |
| chr11_117856643_117857627 | 0.33 |
IL10RA |
interleukin 10 receptor, alpha |
26 |
0.98 |
| chr17_76254452_76254830 | 0.32 |
TMEM235 |
transmembrane protein 235 |
26519 |
0.12 |
| chr16_88714887_88715038 | 0.31 |
CYBA |
cytochrome b-245, alpha polypeptide |
1729 |
0.19 |
| chr4_15661026_15661177 | 0.30 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
386 |
0.87 |
| chr2_43233189_43233340 | 0.29 |
ENSG00000207087 |
. |
85368 |
0.1 |
| chr17_66286056_66286766 | 0.29 |
SLC16A6 |
solute carrier family 16, member 6 |
846 |
0.47 |
| chr19_17862364_17862733 | 0.28 |
FCHO1 |
FCH domain only 1 |
203 |
0.91 |
| chr17_70715496_70715723 | 0.28 |
ENSG00000222545 |
. |
4999 |
0.28 |
| chr17_75868336_75868487 | 0.27 |
FLJ45079 |
|
10248 |
0.24 |
| chr1_28452162_28452313 | 0.26 |
ENSG00000253005 |
. |
17115 |
0.12 |
| chr3_30653355_30653657 | 0.26 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
5413 |
0.33 |
| chrX_12989756_12990021 | 0.26 |
TMSB4X |
thymosin beta 4, X-linked |
3339 |
0.29 |
| chr15_90728173_90728877 | 0.26 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
330 |
0.84 |
| chr17_74072761_74073180 | 0.26 |
GALR2 |
galanin receptor 2 |
2095 |
0.17 |
| chr22_50752570_50752748 | 0.26 |
XX-C283C717.1 |
|
401 |
0.71 |
| chr21_39844669_39844875 | 0.25 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
25573 |
0.26 |
| chr17_33379941_33380092 | 0.25 |
ENSG00000238858 |
. |
1497 |
0.28 |
| chr11_14665333_14666638 | 0.24 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
608 |
0.56 |
| chr17_4487766_4488160 | 0.23 |
SMTNL2 |
smoothelin-like 2 |
129 |
0.94 |
| chr3_190306633_190306784 | 0.23 |
IL1RAP |
interleukin 1 receptor accessory protein |
25473 |
0.22 |
| chr10_73572268_73572419 | 0.23 |
CDH23 |
cadherin-related 23 |
16815 |
0.17 |
| chr20_20692062_20692434 | 0.23 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
883 |
0.69 |
| chr3_53156917_53157068 | 0.22 |
RFT1 |
RFT1 homolog (S. cerevisiae) |
7435 |
0.19 |
| chr17_72008833_72008984 | 0.22 |
RPL38 |
ribosomal protein L38 |
190813 |
0.02 |
| chr8_134088694_134088845 | 0.22 |
SLA |
Src-like-adaptor |
16166 |
0.23 |
| chr12_105044004_105044155 | 0.22 |
ENSG00000264295 |
. |
58668 |
0.13 |
| chr3_172429057_172429256 | 0.22 |
NCEH1 |
neutral cholesterol ester hydrolase 1 |
148 |
0.95 |
| chr11_128591931_128592082 | 0.22 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
26088 |
0.16 |
| chr3_57038204_57038355 | 0.22 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
26876 |
0.18 |
| chr17_79005386_79005537 | 0.22 |
BAIAP2 |
BAI1-associated protein 2 |
3489 |
0.16 |
| chr6_37140965_37141167 | 0.22 |
PIM1 |
pim-1 oncogene |
3087 |
0.24 |
| chr19_51971494_51971730 | 0.21 |
CEACAM18 |
carcinoembryonic antigen-related cell adhesion molecule 18 |
8226 |
0.1 |
| chr19_10405987_10406645 | 0.21 |
ICAM5 |
intercellular adhesion molecule 5, telencephalin |
4663 |
0.08 |
| chr16_88767682_88768006 | 0.21 |
RNF166 |
ring finger protein 166 |
833 |
0.34 |
| chr18_74843262_74844680 | 0.21 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr17_40398832_40398983 | 0.21 |
RP11-358B23.5 |
|
21698 |
0.1 |
| chr17_29821977_29822128 | 0.21 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
6926 |
0.14 |
| chr14_97204377_97204528 | 0.21 |
VRK1 |
vaccinia related kinase 1 |
59189 |
0.14 |
| chr12_66585568_66585719 | 0.20 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
2606 |
0.24 |
| chr17_74261907_74262739 | 0.20 |
UBALD2 |
UBA-like domain containing 2 |
539 |
0.68 |
| chr17_55363036_55363187 | 0.20 |
MSI2 |
musashi RNA-binding protein 2 |
94 |
0.98 |
| chr9_137820763_137820914 | 0.20 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
11029 |
0.14 |
| chr1_37943283_37943786 | 0.20 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
3381 |
0.18 |
| chr16_57721437_57721588 | 0.19 |
CCDC135 |
coiled-coil domain containing 135 |
7193 |
0.13 |
| chr3_43221968_43222122 | 0.19 |
ENSG00000222331 |
. |
30318 |
0.18 |
| chr17_74581180_74582274 | 0.19 |
ST6GALNAC2 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
483 |
0.62 |
| chr1_12533236_12533387 | 0.19 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
5293 |
0.24 |
| chr9_138844390_138844541 | 0.18 |
UBAC1 |
UBA domain containing 1 |
8761 |
0.2 |
| chr13_34116508_34116659 | 0.18 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
191816 |
0.03 |
| chr18_74184609_74184760 | 0.18 |
ZNF516 |
zinc finger protein 516 |
18051 |
0.17 |
| chrX_123105759_123105910 | 0.18 |
STAG2 |
stromal antigen 2 |
8814 |
0.25 |
| chr3_5071207_5071358 | 0.18 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
49636 |
0.12 |
| chr17_75877482_75877763 | 0.18 |
FLJ45079 |
|
1037 |
0.6 |
| chr11_74952584_74952761 | 0.18 |
TPBGL |
trophoblast glycoprotein-like |
722 |
0.6 |
| chr19_44283632_44284486 | 0.18 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
1350 |
0.33 |
| chr15_90608186_90608365 | 0.17 |
ZNF710 |
zinc finger protein 710 |
2971 |
0.2 |
| chr4_166128900_166129705 | 0.17 |
KLHL2 |
kelch-like family member 2 |
327 |
0.55 |
| chr16_53980634_53980785 | 0.17 |
FTO |
fat mass and obesity associated |
12767 |
0.21 |
| chr1_185284734_185285261 | 0.17 |
IVNS1ABP |
influenza virus NS1A binding protein |
1464 |
0.44 |
| chr21_46899502_46899653 | 0.17 |
COL18A1 |
collagen, type XVIII, alpha 1 |
10612 |
0.2 |
| chr15_31507532_31508310 | 0.17 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
54445 |
0.15 |
| chr17_46565994_46566145 | 0.17 |
ENSG00000206805 |
. |
9610 |
0.12 |
| chr2_238263784_238263935 | 0.16 |
COL6A3 |
collagen, type VI, alpha 3 |
32861 |
0.18 |
| chr16_53552293_53552836 | 0.16 |
AKTIP |
AKT interacting protein |
14241 |
0.21 |
| chr15_50399834_50400002 | 0.16 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
6783 |
0.25 |
| chr3_52287768_52287956 | 0.16 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
4885 |
0.1 |
| chr12_54610003_54610573 | 0.16 |
ENSG00000265371 |
. |
14972 |
0.1 |
| chr5_65440502_65441175 | 0.16 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
175 |
0.55 |
| chr9_97402187_97402338 | 0.16 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
157 |
0.97 |
| chr3_14851561_14851984 | 0.15 |
FGD5 |
FYVE, RhoGEF and PH domain containing 5 |
8697 |
0.23 |
| chrX_153209082_153209233 | 0.15 |
RENBP |
renin binding protein |
982 |
0.33 |
| chr22_33223114_33223265 | 0.15 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
25502 |
0.21 |
| chr21_16886585_16887016 | 0.15 |
ENSG00000212564 |
. |
99802 |
0.08 |
| chr20_60760834_60760985 | 0.15 |
MTG2 |
mitochondrial ribosome-associated GTPase 2 |
2802 |
0.2 |
| chr18_24281915_24282066 | 0.15 |
ENSG00000265369 |
. |
12497 |
0.22 |
| chr14_100533458_100533776 | 0.15 |
EVL |
Enah/Vasp-like |
843 |
0.55 |
| chr14_101013926_101014082 | 0.15 |
BEGAIN |
brain-enriched guanylate kinase-associated |
431 |
0.78 |
| chr2_135676446_135677712 | 0.15 |
CCNT2 |
cyclin T2 |
684 |
0.48 |
| chr3_30683174_30683325 | 0.15 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
35156 |
0.21 |
| chr13_28365022_28365726 | 0.15 |
GSX1 |
GS homeobox 1 |
1406 |
0.47 |
| chr12_51402747_51402967 | 0.15 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
175 |
0.93 |
| chr11_48069657_48069808 | 0.14 |
AC103828.1 |
|
32325 |
0.16 |
| chr22_50968790_50969317 | 0.14 |
TYMP |
thymidine phosphorylase |
568 |
0.45 |
| chr1_36838180_36838356 | 0.14 |
STK40 |
serine/threonine kinase 40 |
11327 |
0.12 |
| chr19_19599372_19599523 | 0.14 |
GATAD2A |
GATA zinc finger domain containing 2A |
23157 |
0.1 |
| chr18_11883181_11883332 | 0.14 |
MPPE1 |
metallophosphoesterase 1 |
6158 |
0.13 |
| chrX_95939817_95940209 | 0.14 |
DIAPH2 |
diaphanous-related formin 2 |
302 |
0.93 |
| chr1_182758950_182759312 | 0.14 |
NPL |
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
220 |
0.94 |
| chr21_36280449_36280667 | 0.14 |
RUNX1 |
runt-related transcription factor 1 |
18471 |
0.28 |
| chr19_35609321_35609472 | 0.14 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
38 |
0.95 |
| chr11_65291429_65291724 | 0.14 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
972 |
0.38 |
| chr9_90937776_90937927 | 0.13 |
ENSG00000252299 |
. |
51333 |
0.16 |
| chrX_131623914_131624431 | 0.13 |
MBNL3 |
muscleblind-like splicing regulator 3 |
176 |
0.97 |
| chr2_74063742_74063893 | 0.13 |
STAMBP |
STAM binding protein |
7659 |
0.18 |
| chr4_657472_657717 | 0.13 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
35 |
0.95 |
| chr14_35591199_35591715 | 0.13 |
KIAA0391 |
KIAA0391 |
52 |
0.51 |
| chr14_93672992_93673380 | 0.13 |
C14orf142 |
chromosome 14 open reading frame 142 |
188 |
0.52 |
| chr11_94962772_94964135 | 0.13 |
RP11-712B9.2 |
|
103 |
0.93 |
| chr4_6695738_6695889 | 0.13 |
S100P |
S100 calcium binding protein P |
1017 |
0.4 |
| chr7_142012838_142012989 | 0.13 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
23304 |
0.2 |
| chr7_26952596_26952747 | 0.13 |
SKAP2 |
src kinase associated phosphoprotein 2 |
48309 |
0.14 |
| chr21_45285452_45286244 | 0.13 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
781 |
0.65 |
| chr6_14734934_14735085 | 0.13 |
ENSG00000206960 |
. |
88243 |
0.1 |
| chr15_31650164_31650315 | 0.13 |
KLF13 |
Kruppel-like factor 13 |
8118 |
0.31 |
| chr16_58018530_58018844 | 0.13 |
TEPP |
testis, prostate and placenta expressed |
8348 |
0.13 |
| chr17_27716871_27717334 | 0.12 |
TAOK1 |
TAO kinase 1 |
380 |
0.47 |
| chr5_175560458_175560649 | 0.12 |
FAM153B |
family with sequence similarity 153, member B |
48644 |
0.12 |
| chr6_33545433_33545584 | 0.12 |
BAK1 |
BCL2-antagonist/killer 1 |
2473 |
0.2 |
| chr4_88770427_88770578 | 0.12 |
MEPE |
matrix extracellular phosphoglycoprotein |
16363 |
0.19 |
| chr10_101689512_101689663 | 0.12 |
DNMBP |
dynamin binding protein |
1166 |
0.47 |
| chr9_130666481_130666789 | 0.12 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
974 |
0.33 |
| chr8_95652342_95652927 | 0.12 |
ESRP1 |
epithelial splicing regulatory protein 1 |
668 |
0.52 |
| chr19_2164749_2165707 | 0.12 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
1044 |
0.38 |
| chr7_142902930_142903081 | 0.12 |
TAS2R40 |
taste receptor, type 2, member 40 |
16125 |
0.11 |
| chr2_2690202_2690353 | 0.12 |
ENSG00000263570 |
. |
330825 |
0.01 |
| chr16_56703515_56703666 | 0.12 |
MT1H |
metallothionein 1H |
136 |
0.88 |
| chr4_53525685_53526428 | 0.12 |
USP46 |
ubiquitin specific peptidase 46 |
554 |
0.78 |
| chr7_997543_997694 | 0.12 |
ADAP1 |
ArfGAP with dual PH domains 1 |
2575 |
0.2 |
| chr16_81480914_81481100 | 0.12 |
CMIP |
c-Maf inducing protein |
2232 |
0.39 |
| chr7_142510250_142510438 | 0.11 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
29213 |
0.14 |
| chr11_66311190_66311580 | 0.11 |
ZDHHC24 |
zinc finger, DHHC-type containing 24 |
2321 |
0.15 |
| chr5_156692539_156692693 | 0.11 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
475 |
0.65 |
| chr12_50156324_50156475 | 0.11 |
TMBIM6 |
transmembrane BAX inhibitor motif containing 6 |
9652 |
0.16 |
| chr2_162100897_162101225 | 0.11 |
AC009299.2 |
|
6612 |
0.21 |
| chr6_42018642_42019010 | 0.11 |
TAF8 |
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
164 |
0.87 |
| chr19_34669041_34669419 | 0.11 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
5667 |
0.28 |
| chr2_219085570_219085950 | 0.11 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
3827 |
0.15 |
| chr2_219233529_219233805 | 0.11 |
ENSG00000225062 |
. |
1109 |
0.3 |
| chr17_65417991_65418142 | 0.11 |
ENSG00000201547 |
. |
13069 |
0.14 |
| chr19_10704746_10704897 | 0.11 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
6830 |
0.1 |
| chr13_48893876_48894215 | 0.11 |
RB1 |
retinoblastoma 1 |
16134 |
0.25 |
| chr11_45101502_45101653 | 0.11 |
PRDM11 |
PR domain containing 11 |
13987 |
0.27 |
| chr19_3275241_3275448 | 0.11 |
AC010649.1 |
|
27575 |
0.14 |
| chr7_86946956_86947107 | 0.11 |
CROT |
carnitine O-octanoyltransferase |
27966 |
0.2 |
| chr9_96708088_96708484 | 0.11 |
BARX1 |
BARX homeobox 1 |
7427 |
0.27 |
| chr9_67671299_67671450 | 0.11 |
ENSG00000199432 |
. |
93148 |
0.07 |
| chr15_67316943_67317094 | 0.11 |
SMAD3 |
SMAD family member 3 |
39083 |
0.2 |
| chr8_101964485_101964756 | 0.11 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
160 |
0.95 |
| chr1_38019125_38019899 | 0.11 |
SNIP1 |
Smad nuclear interacting protein 1 |
391 |
0.79 |
| chr22_45625958_45626109 | 0.11 |
KIAA0930 |
KIAA0930 |
3249 |
0.23 |
| chr1_95582420_95583278 | 0.11 |
TMEM56 |
transmembrane protein 56 |
45 |
0.91 |
| chr21_34440256_34440407 | 0.11 |
OLIG1 |
oligodendrocyte transcription factor 1 |
2119 |
0.27 |
| chr13_20795977_20796128 | 0.11 |
GJB6 |
gap junction protein, beta 6, 30kDa |
9065 |
0.2 |
| chr7_156797777_156797998 | 0.11 |
MNX1-AS2 |
MNX1 antisense RNA 2 |
1114 |
0.45 |
| chr9_139925276_139925479 | 0.11 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
2085 |
0.1 |
| chr5_153422227_153422378 | 0.10 |
MFAP3 |
microfibrillar-associated protein 3 |
3743 |
0.24 |
| chr3_45718385_45718593 | 0.10 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
11885 |
0.18 |
| chrX_103357104_103358067 | 0.10 |
ZCCHC18 |
zinc finger, CCHC domain containing 18 |
383 |
0.86 |
| chr3_56949082_56949233 | 0.10 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
1342 |
0.51 |
| chr13_25115593_25116500 | 0.10 |
ENSG00000266216 |
. |
13405 |
0.17 |
| chr16_84631467_84631681 | 0.10 |
RP11-61F12.1 |
|
3575 |
0.21 |
| chr17_25659717_25660000 | 0.10 |
WSB1 |
WD repeat and SOCS box containing 1 |
38486 |
0.16 |
| chr1_182808109_182808399 | 0.10 |
DHX9 |
DEAH (Asp-Glu-Ala-His) box helicase 9 |
250 |
0.93 |
| chr4_16083981_16084260 | 0.10 |
PROM1 |
prominin 1 |
111 |
0.98 |
| chr3_36985452_36986747 | 0.10 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
449 |
0.83 |
| chr17_36859477_36859845 | 0.10 |
ENSG00000265930 |
. |
1077 |
0.24 |
| chr9_123922153_123922304 | 0.10 |
CNTRL |
centriolin |
2107 |
0.3 |
| chr2_118943251_118943914 | 0.10 |
INSIG2 |
insulin induced gene 2 |
97532 |
0.08 |
| chr2_62530766_62530917 | 0.10 |
ENSG00000238809 |
. |
38711 |
0.15 |
| chr9_136207992_136208143 | 0.10 |
ENSG00000201451 |
. |
3406 |
0.08 |
| chr22_43253501_43253661 | 0.10 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
173 |
0.96 |
| chr19_14229110_14229381 | 0.10 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
349 |
0.62 |
| chr17_29814991_29815971 | 0.10 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
355 |
0.83 |
| chr14_78108377_78108627 | 0.10 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
25386 |
0.16 |
| chr12_104572024_104572175 | 0.10 |
TXNRD1 |
thioredoxin reductase 1 |
37458 |
0.13 |
| chr7_65216059_65216706 | 0.10 |
CCT6P1 |
chaperonin containing TCP1, subunit 6 (zeta) pseudogene 1 |
253 |
0.9 |
| chr17_36860259_36860862 | 0.10 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
1235 |
0.2 |
| chr17_19093169_19093320 | 0.10 |
ENSG00000264940 |
. |
314 |
0.77 |
| chr17_29399050_29399201 | 0.10 |
RP11-271K11.5 |
|
21081 |
0.11 |
| chr22_46988879_46989444 | 0.10 |
GRAMD4 |
GRAM domain containing 4 |
16135 |
0.21 |
| chr12_51716816_51717143 | 0.09 |
BIN2 |
bridging integrator 2 |
920 |
0.52 |
| chr9_102584292_102585830 | 0.09 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
924 |
0.68 |
| chr4_57775517_57776061 | 0.09 |
REST |
RE1-silencing transcription factor |
1714 |
0.37 |
| chr16_4357890_4358162 | 0.09 |
GLIS2 |
GLIS family zinc finger 2 |
6736 |
0.14 |
| chr1_203676024_203676175 | 0.09 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
16926 |
0.18 |
| chr14_91125349_91125500 | 0.09 |
TTC7B |
tetratricopeptide repeat domain 7B |
14872 |
0.16 |
| chr16_643128_643307 | 0.09 |
RAB40C |
RAB40C, member RAS oncogene family |
2885 |
0.1 |
| chr8_103251470_103251823 | 0.09 |
KB-431C1.4 |
|
24 |
0.81 |
| chr1_55616949_55617132 | 0.09 |
USP24 |
ubiquitin specific peptidase 24 |
63722 |
0.11 |
| chr9_127531935_127532859 | 0.09 |
RP11-175D17.3 |
|
5 |
0.95 |
| chr20_32700274_32700759 | 0.09 |
EIF2S2 |
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
378 |
0.87 |
| chr3_160473528_160474334 | 0.09 |
PPM1L |
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
65 |
0.95 |
| chr17_1515004_1515155 | 0.09 |
ENSG00000238946 |
. |
4510 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.2 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.4 | GO:1990266 | neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |