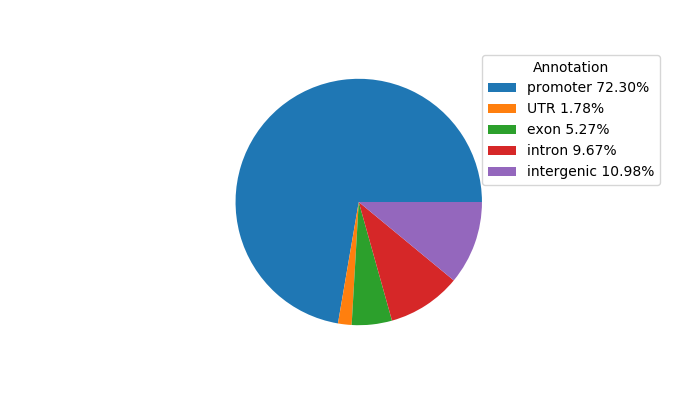
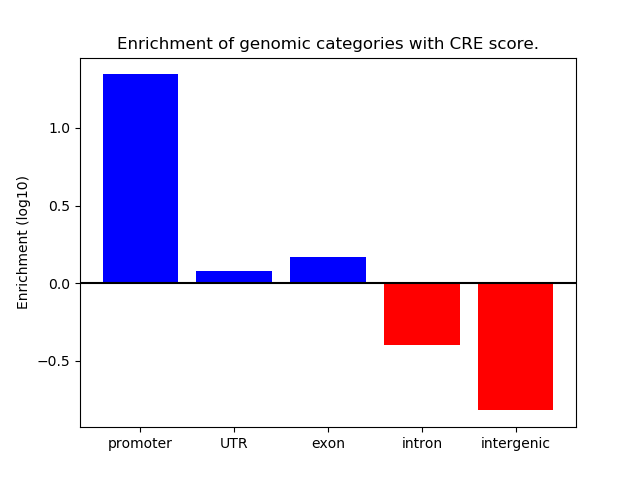
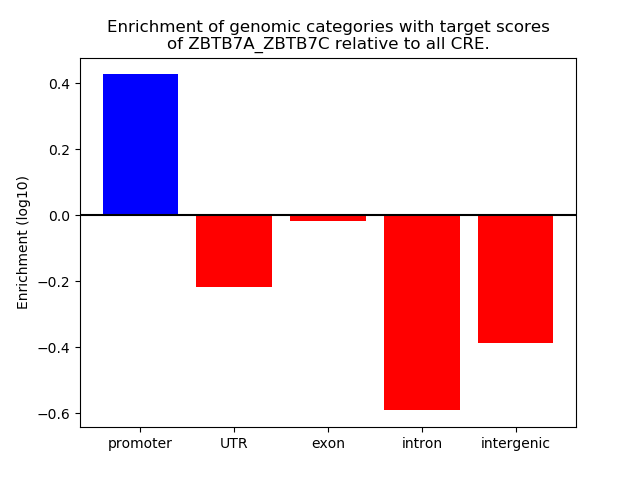
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
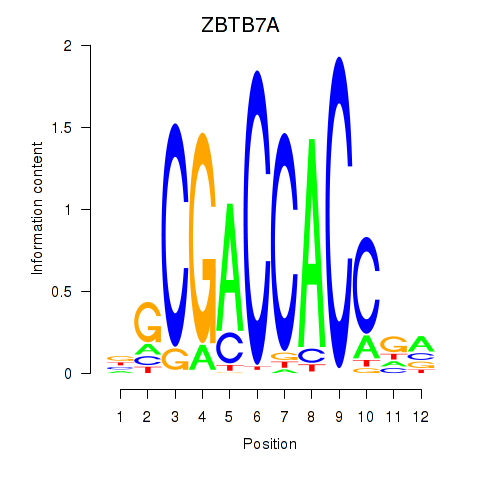
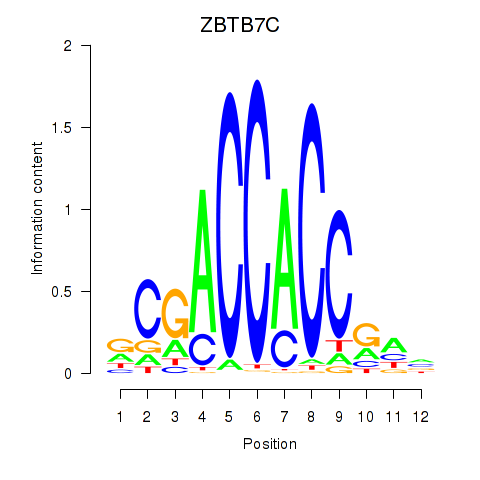
Results for ZBTB7A_ZBTB7C
Z-value: 1.58
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
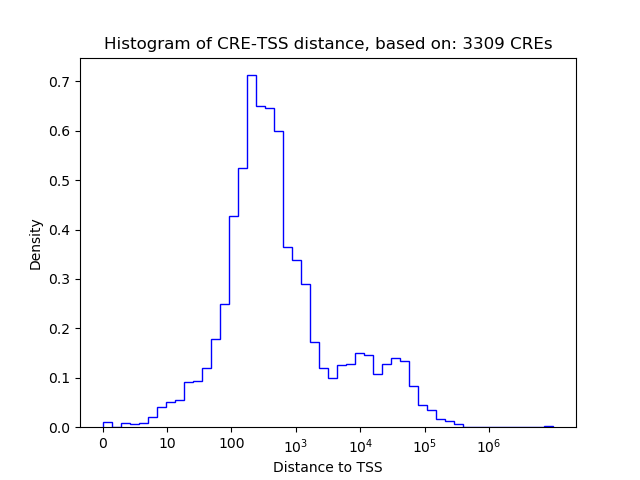
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_4066192_4066601 | ZBTB7A | 547 | 0.639759 | 0.93 | 2.5e-04 | Click! |
| chr19_4064959_4065214 | ZBTB7A | 644 | 0.558006 | 0.89 | 1.2e-03 | Click! |
| chr19_4064037_4064881 | ZBTB7A | 1271 | 0.279646 | 0.83 | 5.8e-03 | Click! |
| chr19_4067333_4067959 | ZBTB7A | 703 | 0.540158 | 0.79 | 1.2e-02 | Click! |
| chr19_4066716_4067152 | ZBTB7A | 9 | 0.959972 | 0.76 | 1.8e-02 | Click! |
| chr18_45968069_45968289 | ZBTB7C | 31056 | 0.165364 | 0.83 | 6.0e-03 | Click! |
| chr18_45534852_45535059 | ZBTB7C | 32539 | 0.210238 | 0.81 | 8.1e-03 | Click! |
| chr18_45534295_45534612 | ZBTB7C | 33041 | 0.208509 | 0.78 | 1.3e-02 | Click! |
| chr18_45662840_45663659 | ZBTB7C | 483 | 0.857360 | 0.75 | 1.9e-02 | Click! |
| chr18_45936037_45936830 | ZBTB7C | 640 | 0.755625 | 0.75 | 2.0e-02 | Click! |
Activity of the ZBTB7A_ZBTB7C motif across conditions
Conditions sorted by the z-value of the ZBTB7A_ZBTB7C motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
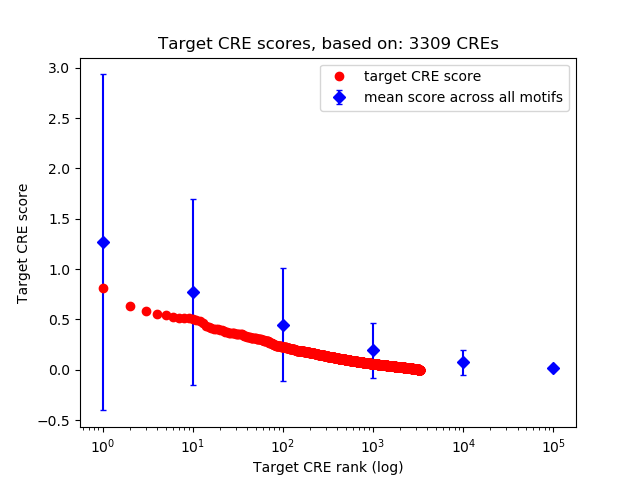
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_145448474_145448628 | 0.82 |
ENSG00000264987 |
. |
7548 |
0.11 |
| chr8_145203218_145203446 | 0.63 |
MROH1 |
maestro heat-like repeat family member 1 |
216 |
0.87 |
| chr15_77712259_77712435 | 0.59 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
95 |
0.84 |
| chr16_89723637_89723841 | 0.56 |
CHMP1A |
charged multivesicular body protein 1A |
319 |
0.57 |
| chr15_62352402_62352620 | 0.54 |
VPS13C |
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
136 |
0.58 |
| chr7_120970338_120970562 | 0.52 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
1393 |
0.55 |
| chr17_42296537_42296906 | 0.52 |
UBTF |
upstream binding transcription factor, RNA polymerase I |
145 |
0.89 |
| chr10_122216781_122217284 | 0.52 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
273 |
0.93 |
| chr15_101458468_101458714 | 0.52 |
RP11-66B24.4 |
|
455 |
0.66 |
| chr8_495850_496251 | 0.50 |
TDRP |
testis development related protein |
269 |
0.95 |
| chr19_1295588_1295739 | 0.50 |
EFNA2 |
ephrin-A2 |
9510 |
0.09 |
| chr11_2398888_2399193 | 0.48 |
ENSG00000238184 |
. |
182 |
0.77 |
| chr4_56212171_56212371 | 0.47 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
5 |
0.98 |
| chr10_95326560_95326711 | 0.44 |
FFAR4 |
free fatty acid receptor 4 |
196 |
0.93 |
| chr7_115994951_115995178 | 0.42 |
ENSG00000216076 |
. |
9066 |
0.21 |
| chr4_779935_780157 | 0.41 |
RP11-440L14.1 |
|
4409 |
0.14 |
| chr22_37730993_37731243 | 0.40 |
CYTH4 |
cytohesin 4 |
25109 |
0.13 |
| chr12_13197333_13197564 | 0.40 |
KIAA1467 |
KIAA1467 |
230 |
0.92 |
| chr11_14912524_14912758 | 0.40 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
556 |
0.81 |
| chr2_32502966_32503192 | 0.40 |
YIPF4 |
Yip1 domain family, member 4 |
100 |
0.97 |
| chr1_20834735_20834945 | 0.39 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
186 |
0.95 |
| chr1_201438046_201438281 | 0.39 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
149 |
0.95 |
| chr20_61492540_61492934 | 0.38 |
TCFL5 |
transcription factor-like 5 (basic helix-loop-helix) |
378 |
0.82 |
| chr5_179779627_179779808 | 0.37 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
183 |
0.96 |
| chr2_96700541_96700692 | 0.37 |
GPAT2 |
glycerol-3-phosphate acyltransferase 2, mitochondrial |
48 |
0.97 |
| chr2_96455429_96455580 | 0.37 |
AC008268.2 |
|
2464 |
0.4 |
| chr11_70962306_70962531 | 0.37 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
1205 |
0.6 |
| chr1_64197464_64197686 | 0.37 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
42118 |
0.17 |
| chr4_72053226_72053895 | 0.36 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
516 |
0.88 |
| chr3_49592272_49592564 | 0.36 |
BSN |
bassoon presynaptic cytomatrix protein |
496 |
0.78 |
| chr17_77966356_77966507 | 0.35 |
CTD-2529O21.1 |
|
209 |
0.88 |
| chr9_91926113_91926303 | 0.35 |
CKS2 |
CDC28 protein kinase regulatory subunit 2 |
95 |
0.94 |
| chr22_45706486_45706804 | 0.35 |
FAM118A |
family with sequence similarity 118, member A |
640 |
0.72 |
| chr2_96874205_96874543 | 0.35 |
STARD7 |
StAR-related lipid transfer (START) domain containing 7 |
182 |
0.54 |
| chr10_49514759_49514955 | 0.35 |
MAPK8 |
mitogen-activated protein kinase 8 |
124 |
0.97 |
| chr2_220306830_220307128 | 0.35 |
SPEG |
SPEG complex locus |
206 |
0.88 |
| chr7_140772581_140772878 | 0.34 |
TMEM178B |
transmembrane protein 178B |
1303 |
0.49 |
| chr17_7492389_7492544 | 0.34 |
SOX15 |
SRY (sex determining region Y)-box 15 |
924 |
0.23 |
| chr7_77167232_77167734 | 0.33 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
97 |
0.98 |
| chr3_13521452_13521618 | 0.33 |
HDAC11-AS1 |
HDAC11 antisense RNA 1 |
18 |
0.63 |
| chr13_110959773_110960124 | 0.33 |
COL4A2 |
collagen, type IV, alpha 2 |
334 |
0.6 |
| chr7_557703_557916 | 0.32 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
336 |
0.89 |
| chr4_17812934_17813275 | 0.32 |
NCAPG |
non-SMC condensin I complex, subunit G |
579 |
0.52 |
| chr2_219264038_219264279 | 0.32 |
RP11-378A13.2 |
|
216 |
0.59 |
| chr13_28366671_28366833 | 0.32 |
GSX1 |
GS homeobox 1 |
28 |
0.98 |
| chr5_158635958_158636135 | 0.32 |
RNF145 |
ring finger protein 145 |
513 |
0.61 |
| chr22_32341496_32341778 | 0.31 |
C22orf24 |
chromosome 22 open reading frame 24 |
301 |
0.81 |
| chr2_241526323_241526743 | 0.31 |
CAPN10 |
calpain 10 |
245 |
0.64 |
| chr6_31619842_31620065 | 0.31 |
BAG6 |
BCL2-associated athanogene 6 |
18 |
0.71 |
| chrX_19905153_19905596 | 0.31 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
203 |
0.96 |
| chr2_151342286_151342580 | 0.31 |
RND3 |
Rho family GTPase 3 |
537 |
0.88 |
| chr1_1819806_1819970 | 0.31 |
GNB1 |
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
1952 |
0.23 |
| chr1_145209374_145209525 | 0.31 |
NOTCH2NL |
notch 2 N-terminal like |
330 |
0.9 |
| chr7_1497220_1497371 | 0.31 |
MICALL2 |
MICAL-like 2 |
1667 |
0.29 |
| chr17_75084804_75085699 | 0.30 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
14 |
0.64 |
| chr17_36903993_36904433 | 0.30 |
PCGF2 |
polycomb group ring finger 2 |
224 |
0.79 |
| chr14_93650615_93650845 | 0.30 |
MOAP1 |
modulator of apoptosis 1 |
538 |
0.33 |
| chr17_79361447_79361695 | 0.30 |
RP11-1055B8.6 |
Uncharacterized protein |
7704 |
0.12 |
| chr11_129872200_129872372 | 0.30 |
PRDM10 |
PR domain containing 10 |
394 |
0.89 |
| chr22_38350140_38350321 | 0.29 |
POLR2F |
polymerase (RNA) II (DNA directed) polypeptide F |
496 |
0.43 |
| chr9_135545828_135545979 | 0.29 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
115 |
0.85 |
| chr3_36422153_36422411 | 0.29 |
STAC |
SH3 and cysteine rich domain |
217 |
0.97 |
| chr1_210406979_210407193 | 0.29 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
303 |
0.83 |
| chr7_117854626_117855054 | 0.29 |
ANKRD7 |
ankyrin repeat domain 7 |
9872 |
0.26 |
| chr22_18483386_18483604 | 0.29 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
451 |
0.77 |
| chr19_56879183_56879365 | 0.29 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
478 |
0.62 |
| chr19_3985625_3985776 | 0.29 |
EEF2 |
eukaryotic translation elongation factor 2 |
233 |
0.86 |
| chr22_35654352_35654529 | 0.28 |
HMGXB4 |
HMG box domain containing 4 |
949 |
0.54 |
| chr9_33818545_33819028 | 0.28 |
RP11-133O22.6 |
|
7 |
0.95 |
| chr6_16760801_16761048 | 0.27 |
RP1-151F17.1 |
|
445 |
0.67 |
| chr6_160210417_160210645 | 0.27 |
TCP1 |
t-complex 1 |
73 |
0.92 |
| chr11_1330505_1330784 | 0.27 |
TOLLIP |
toll interacting protein |
205 |
0.66 |
| chr1_1284213_1284419 | 0.26 |
DVL1 |
dishevelled segment polarity protein 1 |
176 |
0.85 |
| chr1_183605727_183605878 | 0.26 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
558 |
0.58 |
| chr1_161419213_161419364 | 0.26 |
ENSG00000206921 |
. |
48209 |
0.09 |
| chr19_4402510_4402661 | 0.26 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
74 |
0.93 |
| chr7_73704189_73704652 | 0.26 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
615 |
0.73 |
| chr17_47840525_47840864 | 0.26 |
FAM117A |
family with sequence similarity 117, member A |
799 |
0.59 |
| chr2_217559691_217559970 | 0.25 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
313 |
0.72 |
| chr12_108523289_108523785 | 0.25 |
WSCD2 |
WSC domain containing 2 |
26 |
0.99 |
| chr8_681413_681602 | 0.25 |
ERICH1 |
glutamate-rich 1 |
268 |
0.92 |
| chr2_242499516_242499768 | 0.24 |
BOK-AS1 |
BOK antisense RNA 1 |
1250 |
0.3 |
| chr6_28602550_28602995 | 0.24 |
ENSG00000272278 |
. |
13270 |
0.2 |
| chr5_56247782_56247935 | 0.24 |
MIER3 |
mesoderm induction early response 1, family member 3 |
82 |
0.96 |
| chr7_1609792_1610072 | 0.24 |
PSMG3 |
proteasome (prosome, macropain) assembly chaperone 3 |
251 |
0.91 |
| chr9_131450959_131451258 | 0.24 |
SET |
SET nuclear oncogene |
397 |
0.73 |
| chr15_91475533_91475745 | 0.24 |
HDDC3 |
HD domain containing 3 |
67 |
0.93 |
| chr9_108456841_108457052 | 0.24 |
TMEM38B |
transmembrane protein 38B |
121 |
0.97 |
| chr3_197476356_197476565 | 0.24 |
FYTTD1 |
forty-two-three domain containing 1 |
36 |
0.58 |
| chr14_105830576_105830747 | 0.24 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
18795 |
0.13 |
| chr20_21495239_21495416 | 0.24 |
NKX2-2 |
NK2 homeobox 2 |
663 |
0.72 |
| chr10_70091844_70092097 | 0.24 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
91 |
0.91 |
| chr22_21058522_21058673 | 0.24 |
POM121L4P |
POM121 transmembrane nucleoporin-like 4 pseudogene |
14380 |
0.13 |
| chr19_531951_532235 | 0.23 |
CDC34 |
cell division cycle 34 |
33 |
0.95 |
| chr8_6566214_6566531 | 0.23 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
158 |
0.87 |
| chr16_21313256_21313440 | 0.23 |
CRYM |
crystallin, mu |
1024 |
0.57 |
| chr1_161434016_161434167 | 0.23 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
41129 |
0.1 |
| chr1_2518255_2518458 | 0.23 |
FAM213B |
family with sequence similarity 213, member B |
40 |
0.95 |
| chr17_27044345_27044901 | 0.23 |
RAB34 |
RAB34, member RAS oncogene family |
143 |
0.82 |
| chr10_5855093_5855330 | 0.23 |
GDI2 |
GDP dissociation inhibitor 2 |
139 |
0.96 |
| chr5_98109376_98109557 | 0.23 |
RGMB |
repulsive guidance molecule family member b |
127 |
0.72 |
| chr4_54930355_54930595 | 0.23 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
8 |
0.97 |
| chr8_145010051_145010202 | 0.23 |
PLEC |
plectin |
3632 |
0.14 |
| chr12_120966627_120966937 | 0.22 |
COQ5 |
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
161 |
0.91 |
| chr20_62152329_62152480 | 0.22 |
PPDPF |
pancreatic progenitor cell differentiation and proliferation factor |
276 |
0.83 |
| chr4_2933657_2933808 | 0.22 |
MFSD10 |
major facilitator superfamily domain containing 10 |
1942 |
0.24 |
| chr11_110168583_110168734 | 0.22 |
RDX |
radixin |
1211 |
0.64 |
| chr8_101965049_101965275 | 0.22 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
49 |
0.97 |
| chr17_80170261_80170703 | 0.22 |
CCDC57 |
coiled-coil domain containing 57 |
207 |
0.87 |
| chr1_214723918_214724069 | 0.22 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
573 |
0.84 |
| chr22_19466002_19466444 | 0.22 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
231 |
0.79 |
| chr5_9545673_9545824 | 0.22 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
439 |
0.82 |
| chr9_102058504_102058983 | 0.22 |
ENSG00000222337 |
. |
12439 |
0.24 |
| chr16_31885188_31885435 | 0.22 |
ZNF267 |
zinc finger protein 267 |
76 |
0.98 |
| chr3_136471001_136471256 | 0.22 |
STAG1 |
stromal antigen 1 |
80 |
0.81 |
| chr5_107005011_107005570 | 0.22 |
EFNA5 |
ephrin-A5 |
1038 |
0.67 |
| chr15_42119664_42119815 | 0.22 |
RP11-23P13.4 |
|
314 |
0.43 |
| chr1_154531335_154531531 | 0.21 |
UBE2Q1 |
ubiquitin-conjugating enzyme E2Q family member 1 |
324 |
0.81 |
| chr2_113239211_113239607 | 0.21 |
TTL |
tubulin tyrosine ligase |
322 |
0.9 |
| chr21_27011889_27012538 | 0.21 |
JAM2 |
junctional adhesion molecule 2 |
303 |
0.91 |
| chr11_64527075_64527226 | 0.21 |
PYGM |
phosphorylase, glycogen, muscle |
296 |
0.84 |
| chr18_32556796_32556947 | 0.21 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
21 |
0.99 |
| chr12_32832317_32832517 | 0.21 |
DNM1L |
dynamin 1-like |
147 |
0.97 |
| chr3_111578032_111578464 | 0.21 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
110 |
0.98 |
| chr2_44396060_44396215 | 0.21 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
88 |
0.57 |
| chr14_68283028_68283289 | 0.21 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
146 |
0.95 |
| chr17_72732981_72733167 | 0.21 |
RAB37 |
RAB37, member RAS oncogene family |
18 |
0.95 |
| chr10_70940505_70940781 | 0.20 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
599 |
0.72 |
| chr19_35491304_35491470 | 0.20 |
GRAMD1A |
GRAM domain containing 1A |
27 |
0.81 |
| chr9_117267117_117267337 | 0.20 |
DFNB31 |
deafness, autosomal recessive 31 |
23 |
0.98 |
| chr15_83876664_83877141 | 0.20 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
132 |
0.96 |
| chr7_77168081_77168577 | 0.20 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
943 |
0.69 |
| chr16_83841657_83841832 | 0.20 |
HSBP1 |
heat shock factor binding protein 1 |
150 |
0.7 |
| chr3_196044847_196045072 | 0.20 |
TCTEX1D2 |
Tctex1 domain containing 2 |
200 |
0.53 |
| chr2_175201680_175202081 | 0.20 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
271 |
0.89 |
| chr22_42321871_42322361 | 0.20 |
TNFRSF13C |
tumor necrosis factor receptor superfamily, member 13C |
706 |
0.48 |
| chr17_21117695_21117937 | 0.20 |
TMEM11 |
transmembrane protein 11 |
121 |
0.57 |
| chr18_9017410_9017607 | 0.20 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
85120 |
0.08 |
| chr3_123753181_123753332 | 0.20 |
ENSG00000238512 |
. |
27675 |
0.16 |
| chr3_9833920_9834114 | 0.20 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
162 |
0.69 |
| chr3_100119741_100120229 | 0.19 |
LNP1 |
leukemia NUP98 fusion partner 1 |
52 |
0.77 |
| chr1_161441388_161441539 | 0.19 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
33757 |
0.12 |
| chr1_182992581_182992854 | 0.19 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
122 |
0.97 |
| chr6_144163877_144164309 | 0.19 |
LTV1 |
LTV1 homolog (S. cerevisiae) |
388 |
0.89 |
| chr9_132359092_132359260 | 0.19 |
ENSG00000216181 |
. |
5335 |
0.14 |
| chr19_801774_802209 | 0.19 |
PTBP1 |
polypyrimidine tract binding protein 1 |
2632 |
0.12 |
| chr9_108320464_108320814 | 0.19 |
FKTN |
fukutin |
226 |
0.62 |
| chr17_40834675_40835133 | 0.19 |
CCR10 |
chemokine (C-C motif) receptor 10 |
178 |
0.59 |
| chr5_172754839_172755250 | 0.19 |
STC2 |
stanniocalcin 2 |
12 |
0.98 |
| chr11_118978157_118978348 | 0.19 |
C2CD2L |
C2CD2-like |
159 |
0.82 |
| chr14_55657735_55658354 | 0.19 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
154 |
0.97 |
| chr12_52890104_52890255 | 0.19 |
KRT6A |
keratin 6A |
3138 |
0.14 |
| chr4_7045813_7046021 | 0.19 |
TADA2B |
transcriptional adaptor 2B |
655 |
0.47 |
| chr16_28874872_28875091 | 0.19 |
SH2B1 |
SH2B adaptor protein 1 |
101 |
0.74 |
| chr5_89825640_89826074 | 0.19 |
LYSMD3 |
LysM, putative peptidoglycan-binding, domain containing 3 |
456 |
0.61 |
| chr4_2819558_2819838 | 0.19 |
SH3BP2 |
SH3-domain binding protein 2 |
214 |
0.94 |
| chr13_41239296_41239826 | 0.19 |
FOXO1 |
forkhead box O1 |
1173 |
0.56 |
| chr12_107349405_107349556 | 0.19 |
C12orf23 |
chromosome 12 open reading frame 23 |
17 |
0.73 |
| chr10_51827733_51827958 | 0.19 |
FAM21A |
family with sequence similarity 21, member A |
124 |
0.97 |
| chr17_78120589_78120769 | 0.19 |
EIF4A3 |
eukaryotic translation initiation factor 4A3 |
259 |
0.9 |
| chr13_41364266_41364578 | 0.19 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
789 |
0.61 |
| chr9_139333187_139333496 | 0.19 |
INPP5E |
inositol polyphosphate-5-phosphatase, 72 kDa |
933 |
0.4 |
| chr1_51702634_51703015 | 0.19 |
RNF11 |
ring finger protein 11 |
881 |
0.51 |
| chr1_6674083_6674241 | 0.18 |
PHF13 |
PHD finger protein 13 |
417 |
0.47 |
| chr1_161440532_161440683 | 0.18 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
34613 |
0.12 |
| chr3_133646028_133646432 | 0.18 |
C3orf36 |
chromosome 3 open reading frame 36 |
2426 |
0.34 |
| chr19_10443271_10443899 | 0.18 |
RAVER1 |
ribonucleoprotein, PTB-binding 1 |
731 |
0.41 |
| chr2_677442_677606 | 0.18 |
TMEM18 |
transmembrane protein 18 |
85 |
0.8 |
| chr8_145703691_145703928 | 0.18 |
FOXH1 |
forkhead box H1 |
2091 |
0.11 |
| chr12_49183193_49183344 | 0.18 |
ADCY6 |
adenylate cyclase 6 |
448 |
0.69 |
| chr7_91875452_91875672 | 0.18 |
ANKIB1 |
ankyrin repeat and IBR domain containing 1 |
14 |
0.55 |
| chr2_132182935_132183127 | 0.18 |
TUBA3D |
tubulin, alpha 3d |
50635 |
0.12 |
| chr9_86432368_86432590 | 0.18 |
GKAP1 |
G kinase anchoring protein 1 |
68 |
0.97 |
| chr18_19321520_19322173 | 0.18 |
MIB1 |
mindbomb E3 ubiquitin protein ligase 1 |
565 |
0.64 |
| chr7_72971746_72971941 | 0.18 |
BCL7B |
B-cell CLL/lymphoma 7B |
158 |
0.94 |
| chr20_60757985_60758272 | 0.18 |
MTG2 |
mitochondrial ribosome-associated GTPase 2 |
21 |
0.97 |
| chr17_3438791_3438956 | 0.18 |
SPATA22 |
spermatogenesis associated 22 |
21727 |
0.11 |
| chr13_23489998_23490201 | 0.18 |
ENSG00000240341 |
. |
36761 |
0.19 |
| chr1_1823304_1823560 | 0.18 |
RP1-140A9.1 |
|
522 |
0.58 |
| chr17_20059381_20059604 | 0.18 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
90 |
0.98 |
| chr5_110559444_110559831 | 0.18 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
11 |
0.98 |
| chr11_76495074_76495334 | 0.18 |
TSKU |
tsukushi, small leucine rich proteoglycan |
12 |
0.95 |
| chr10_35928586_35929142 | 0.18 |
ENSG00000264780 |
. |
1316 |
0.35 |
| chr8_128747670_128748110 | 0.18 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
125 |
0.98 |
| chr2_219824025_219824230 | 0.18 |
CDK5R2 |
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
250 |
0.84 |
| chr9_132935193_132935377 | 0.18 |
NCS1 |
neuronal calcium sensor 1 |
428 |
0.85 |
| chr14_96000920_96001139 | 0.17 |
GLRX5 |
glutaredoxin 5 |
99 |
0.49 |
| chr8_55014181_55014390 | 0.17 |
LYPLA1 |
lysophospholipase I |
103 |
0.97 |
| chr9_100149677_100149828 | 0.17 |
ENSG00000221269 |
. |
23789 |
0.18 |
| chr5_110559928_110560116 | 0.17 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
238 |
0.95 |
| chr15_92397216_92397409 | 0.17 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
39 |
0.98 |
| chr19_44951986_44952721 | 0.17 |
ZNF229 |
zinc finger protein 229 |
296 |
0.89 |
| chr22_42949413_42949804 | 0.17 |
SERHL2 |
serine hydrolase-like 2 |
15 |
0.95 |
| chr11_61314245_61314396 | 0.17 |
SYT7 |
synaptotagmin VII |
33972 |
0.11 |
| chr7_99679166_99679374 | 0.17 |
ZNF3 |
zinc finger protein 3 |
58 |
0.92 |
| chr15_85113701_85114038 | 0.17 |
UBE2Q2P1 |
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 1 |
62 |
0.77 |
| chr12_122985185_122985426 | 0.17 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
163 |
0.94 |
| chr2_90413521_90413837 | 0.17 |
CH17-132F21.1 |
Uncharacterized protein |
44522 |
0.15 |
| chr2_205410277_205410441 | 0.17 |
PARD3B |
par-3 family cell polarity regulator beta |
157 |
0.98 |
| chr21_18985375_18985638 | 0.17 |
BTG3 |
BTG family, member 3 |
241 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.4 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.1 | 0.3 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.3 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 | 0.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.2 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.2 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.1 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 0.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.1 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.1 | 0.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 0.1 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.1 | 0.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0051955 | regulation of amino acid transport(GO:0051955) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0033240 | positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.0 | 0.0 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0042161 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:1903019 | negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle organ development(GO:0048635) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.0 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 1.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.0 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0014047 | glutamate secretion(GO:0014047) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.4 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.1 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.4 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.0 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.1 | 0.4 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.1 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.4 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.2 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |