Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
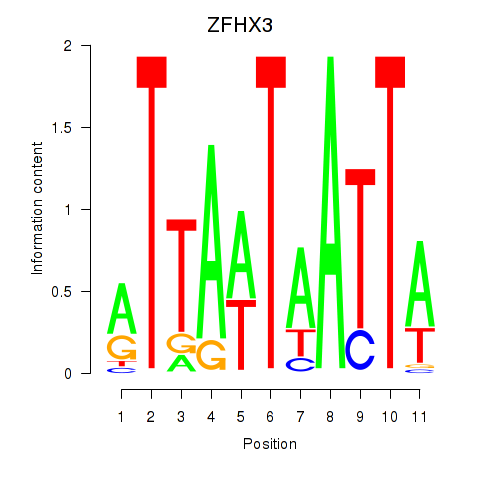
Results for ZFHX3
Z-value: 1.23
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | zinc finger homeobox 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_73088507_73088710 | ZFHX3 | 4989 | 0.255734 | -0.87 | 2.2e-03 | Click! |
| chr16_73086572_73086723 | ZFHX3 | 4373 | 0.262989 | -0.80 | 8.9e-03 | Click! |
| chr16_73105015_73105287 | ZFHX3 | 11554 | 0.234841 | -0.76 | 1.7e-02 | Click! |
| chr16_73102617_73102909 | ZFHX3 | 9166 | 0.240506 | -0.72 | 2.8e-02 | Click! |
| chr16_73093289_73093448 | ZFHX3 | 229 | 0.948003 | -0.72 | 2.8e-02 | Click! |
Activity of the ZFHX3 motif across conditions
Conditions sorted by the z-value of the ZFHX3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
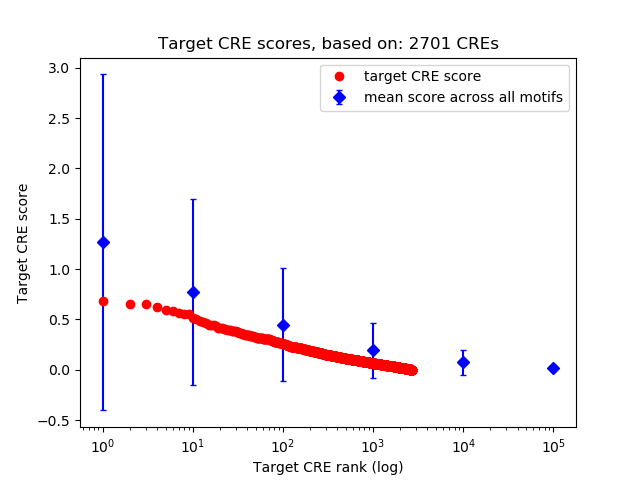
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_20336872_20337023 | 0.68 |
E2F3 |
E2F transcription factor 3 |
65451 |
0.1 |
| chr8_134086469_134086645 | 0.66 |
SLA |
Src-like-adaptor |
13954 |
0.23 |
| chr2_61111507_61111691 | 0.66 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
2808 |
0.27 |
| chr7_150414678_150414897 | 0.63 |
GIMAP1 |
GTPase, IMAP family member 1 |
1142 |
0.45 |
| chr8_126960676_126960849 | 0.59 |
ENSG00000206695 |
. |
47567 |
0.2 |
| chr5_40686354_40686614 | 0.58 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
6884 |
0.22 |
| chrX_78400540_78401325 | 0.56 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chrX_146273220_146273371 | 0.55 |
ENSG00000216171 |
. |
1990 |
0.19 |
| chrX_84258496_84258805 | 0.55 |
APOOL |
apolipoprotein O-like |
182 |
0.97 |
| chrX_22878429_22878580 | 0.52 |
DDX53 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 |
139583 |
0.05 |
| chr3_24225373_24225524 | 0.51 |
THRB |
thyroid hormone receptor, beta |
18362 |
0.28 |
| chr14_22968352_22968523 | 0.48 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
12266 |
0.1 |
| chr15_60883034_60883482 | 0.47 |
RORA |
RAR-related orphan receptor A |
1482 |
0.47 |
| chrX_37910743_37910894 | 0.47 |
SYTL5 |
synaptotagmin-like 5 |
18031 |
0.2 |
| chr5_67576888_67577103 | 0.45 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
860 |
0.74 |
| chr9_33446798_33447442 | 0.44 |
AQP3 |
aquaporin 3 (Gill blood group) |
489 |
0.76 |
| chr4_122148073_122148224 | 0.44 |
TNIP3 |
TNFAIP3 interacting protein 3 |
473 |
0.85 |
| chr5_119523820_119523971 | 0.43 |
ENSG00000251975 |
. |
149453 |
0.05 |
| chr9_134151798_134152108 | 0.42 |
FAM78A |
family with sequence similarity 78, member A |
19 |
0.98 |
| chr7_142430842_142430993 | 0.42 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
26402 |
0.17 |
| chr14_71455264_71455741 | 0.42 |
PCNX |
pecanex homolog (Drosophila) |
24246 |
0.26 |
| chr19_54465805_54465956 | 0.40 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
414 |
0.72 |
| chr13_41540610_41540946 | 0.40 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
15640 |
0.18 |
| chr14_100689805_100690076 | 0.40 |
YY1 |
YY1 transcription factor |
14695 |
0.11 |
| chr9_34745557_34745708 | 0.39 |
FAM205A |
family with sequence similarity 205, member A |
16168 |
0.13 |
| chr1_113164228_113164419 | 0.39 |
ST7L |
suppression of tumorigenicity 7 like |
2283 |
0.19 |
| chr14_22947593_22947744 | 0.38 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr1_200865204_200865355 | 0.38 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr1_247333614_247333765 | 0.38 |
ZNF124 |
zinc finger protein 124 |
1580 |
0.32 |
| chr11_89271457_89271608 | 0.38 |
NOX4 |
NADPH oxidase 4 |
40177 |
0.2 |
| chr1_33447039_33447276 | 0.38 |
RP1-117O3.2 |
|
5519 |
0.15 |
| chr12_22565647_22565798 | 0.37 |
C2CD5 |
C2 calcium-dependent domain containing 5 |
59517 |
0.14 |
| chr4_48134207_48134444 | 0.37 |
TXK |
TXK tyrosine kinase |
1948 |
0.29 |
| chr11_118213949_118214100 | 0.36 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
565 |
0.54 |
| chr6_128221212_128221624 | 0.36 |
THEMIS |
thymocyte selection associated |
685 |
0.81 |
| chr1_25889724_25889930 | 0.36 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
19756 |
0.18 |
| chr2_191225245_191225396 | 0.36 |
INPP1 |
inositol polyphosphate-1-phosphatase |
4058 |
0.23 |
| chr8_115225029_115225180 | 0.35 |
ENSG00000206719 |
. |
261989 |
0.02 |
| chr3_33061626_33061777 | 0.35 |
CCR4 |
chemokine (C-C motif) receptor 4 |
68635 |
0.09 |
| chr6_24929304_24929455 | 0.34 |
FAM65B |
family with sequence similarity 65, member B |
6809 |
0.24 |
| chr18_56326442_56326593 | 0.34 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
12101 |
0.14 |
| chr8_17782406_17782557 | 0.34 |
PCM1 |
pericentriolar material 1 |
228 |
0.93 |
| chr11_95420136_95420287 | 0.34 |
FAM76B |
family with sequence similarity 76, member B |
99758 |
0.08 |
| chr12_47611625_47611801 | 0.34 |
PCED1B |
PC-esterase domain containing 1B |
1332 |
0.49 |
| chr12_69724195_69724437 | 0.34 |
LYZ |
lysozyme |
17805 |
0.16 |
| chr6_106969102_106969411 | 0.33 |
AIM1 |
absent in melanoma 1 |
9526 |
0.21 |
| chr12_76271082_76271233 | 0.33 |
ENSG00000243420 |
. |
80753 |
0.1 |
| chr11_128387324_128387802 | 0.32 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4533 |
0.26 |
| chr17_29645805_29646348 | 0.32 |
CTD-2370N5.3 |
|
227 |
0.88 |
| chr1_158903283_158903483 | 0.32 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
2025 |
0.36 |
| chr17_75453810_75454179 | 0.32 |
SEPT9 |
septin 9 |
1546 |
0.33 |
| chr5_39203281_39203432 | 0.32 |
FYB |
FYN binding protein |
227 |
0.96 |
| chr19_4535398_4535574 | 0.31 |
PLIN5 |
perilipin 5 |
250 |
0.84 |
| chr20_18446613_18446764 | 0.31 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
663 |
0.48 |
| chr1_160611595_160611756 | 0.31 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
5136 |
0.18 |
| chr6_128239129_128239316 | 0.31 |
THEMIS |
thymocyte selection associated |
463 |
0.89 |
| chr6_26198173_26198564 | 0.31 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr1_65428751_65428968 | 0.31 |
JAK1 |
Janus kinase 1 |
3328 |
0.31 |
| chr3_16347363_16347514 | 0.31 |
RP11-415F23.2 |
|
8508 |
0.17 |
| chr9_135038389_135038692 | 0.31 |
NTNG2 |
netrin G2 |
1206 |
0.56 |
| chr3_66549418_66550156 | 0.31 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
1569 |
0.56 |
| chr20_43597999_43598419 | 0.31 |
STK4 |
serine/threonine kinase 4 |
3042 |
0.19 |
| chr1_24862040_24862255 | 0.31 |
ENSG00000266551 |
. |
5943 |
0.18 |
| chrX_78621479_78621720 | 0.31 |
ITM2A |
integral membrane protein 2A |
1257 |
0.64 |
| chr1_38461172_38461323 | 0.31 |
ENSG00000212541 |
. |
4113 |
0.14 |
| chr3_46413606_46413757 | 0.31 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
480 |
0.77 |
| chr12_118796743_118797337 | 0.31 |
TAOK3 |
TAO kinase 3 |
69 |
0.98 |
| chr1_100573281_100573432 | 0.31 |
RP4-714D9.2 |
|
23329 |
0.12 |
| chr13_69182775_69182926 | 0.31 |
ENSG00000243671 |
. |
85363 |
0.11 |
| chr18_34410889_34411040 | 0.30 |
KIAA1328 |
KIAA1328 |
1673 |
0.34 |
| chr16_4538279_4538430 | 0.30 |
HMOX2 |
heme oxygenase (decycling) 2 |
7505 |
0.13 |
| chr19_2819518_2819756 | 0.29 |
ZNF554 |
zinc finger protein 554 |
235 |
0.87 |
| chr13_41553714_41553865 | 0.29 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
2629 |
0.29 |
| chr1_7946262_7946413 | 0.29 |
UTS2 |
urotensin 2 |
32765 |
0.14 |
| chr6_139352039_139352190 | 0.29 |
ABRACL |
ABRA C-terminal like |
2135 |
0.38 |
| chr12_8240325_8240677 | 0.29 |
NECAP1 |
NECAP endocytosis associated 1 |
5118 |
0.16 |
| chr2_197127548_197127913 | 0.29 |
AC020571.3 |
|
2573 |
0.29 |
| chr2_55236798_55237891 | 0.29 |
RTN4 |
reticulon 4 |
241 |
0.93 |
| chr3_141249287_141249763 | 0.29 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
5550 |
0.26 |
| chr2_242051303_242051464 | 0.28 |
MTERFD2 |
MTERF domain containing 2 |
9636 |
0.14 |
| chr6_144668099_144668390 | 0.28 |
UTRN |
utrophin |
3007 |
0.32 |
| chr1_206731858_206732141 | 0.28 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
1506 |
0.37 |
| chr12_18841597_18841748 | 0.28 |
PLCZ1 |
phospholipase C, zeta 1 |
6299 |
0.25 |
| chr5_156609776_156609927 | 0.28 |
ITK |
IL2-inducible T-cell kinase |
2014 |
0.23 |
| chr11_124665951_124666102 | 0.28 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
4253 |
0.12 |
| chr6_154570961_154571273 | 0.28 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
2301 |
0.45 |
| chr10_17548651_17548924 | 0.28 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
52458 |
0.13 |
| chr8_82020312_82020466 | 0.27 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
3914 |
0.34 |
| chr4_90220151_90220996 | 0.27 |
GPRIN3 |
GPRIN family member 3 |
8588 |
0.31 |
| chr10_8099034_8099185 | 0.27 |
GATA3 |
GATA binding protein 3 |
2340 |
0.45 |
| chr18_12847485_12847636 | 0.27 |
PTPN2 |
protein tyrosine phosphatase, non-receptor type 2 |
3580 |
0.29 |
| chr10_119891870_119892021 | 0.27 |
CASC2 |
cancer susceptibility candidate 2 (non-protein coding) |
82820 |
0.09 |
| chr4_93156215_93156366 | 0.27 |
RP11-9B6.1 |
HCG2045747; Uncharacterized protein |
69039 |
0.13 |
| chr13_46755773_46755938 | 0.27 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
604 |
0.7 |
| chr8_100795778_100795929 | 0.27 |
RP11-402L5.1 |
|
15562 |
0.25 |
| chr16_89551290_89551568 | 0.27 |
ANKRD11 |
ankyrin repeat domain 11 |
5297 |
0.13 |
| chr1_192547268_192547419 | 0.26 |
RGS1 |
regulator of G-protein signaling 1 |
2440 |
0.31 |
| chrX_84632426_84632577 | 0.26 |
POF1B |
premature ovarian failure, 1B |
2221 |
0.45 |
| chr1_109188702_109189117 | 0.26 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
14768 |
0.19 |
| chrX_118926003_118926336 | 0.26 |
RPL39 |
ribosomal protein L39 |
563 |
0.7 |
| chr8_38243294_38243670 | 0.26 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
243 |
0.88 |
| chr3_151917198_151917573 | 0.26 |
MBNL1 |
muscleblind-like splicing regulator 1 |
68444 |
0.12 |
| chr11_47787507_47787658 | 0.26 |
FNBP4 |
formin binding protein 4 |
1271 |
0.36 |
| chr1_84611671_84611822 | 0.25 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
1792 |
0.48 |
| chr7_35731550_35731936 | 0.25 |
HERPUD2 |
HERPUD family member 2 |
2433 |
0.31 |
| chrY_8388065_8388216 | 0.25 |
AC007967.3 |
|
386125 |
0.01 |
| chr6_149729706_149729857 | 0.25 |
SUMO4 |
small ubiquitin-like modifier 4 |
8286 |
0.19 |
| chr2_31547763_31547914 | 0.25 |
XDH |
xanthine dehydrogenase |
89743 |
0.08 |
| chr11_128385412_128385563 | 0.25 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6609 |
0.25 |
| chr11_16946345_16946496 | 0.25 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
43515 |
0.12 |
| chr3_63954713_63955178 | 0.25 |
ATXN7 |
ataxin 7 |
1525 |
0.34 |
| chr7_138788856_138789199 | 0.25 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
5073 |
0.23 |
| chr2_223290336_223290619 | 0.24 |
SGPP2 |
sphingosine-1-phosphate phosphatase 2 |
1241 |
0.5 |
| chr4_109086470_109087224 | 0.24 |
LEF1 |
lymphoid enhancer-binding factor 1 |
610 |
0.72 |
| chr9_36152272_36152423 | 0.24 |
GLIPR2 |
GLI pathogenesis-related 2 |
15605 |
0.17 |
| chr1_198654198_198654380 | 0.24 |
RP11-553K8.5 |
|
18099 |
0.24 |
| chr8_52771885_52772036 | 0.24 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
13655 |
0.18 |
| chr1_116293768_116293919 | 0.24 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
17492 |
0.24 |
| chr3_168234505_168234656 | 0.24 |
ENSG00000207717 |
. |
35062 |
0.2 |
| chr5_40678072_40678223 | 0.23 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
1453 |
0.44 |
| chr7_38343163_38343314 | 0.23 |
STARD3NL |
STARD3 N-terminal like |
125241 |
0.06 |
| chr17_47766049_47766200 | 0.23 |
SPOP |
speckle-type POZ protein |
10528 |
0.14 |
| chr3_186743992_186744192 | 0.23 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
821 |
0.7 |
| chr8_82022969_82023806 | 0.23 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
916 |
0.72 |
| chr3_29395618_29395769 | 0.23 |
ENSG00000216169 |
. |
15219 |
0.25 |
| chr11_2405249_2405453 | 0.23 |
CD81 |
CD81 molecule |
529 |
0.65 |
| chr8_8706094_8706267 | 0.23 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
44975 |
0.14 |
| chr5_118606441_118606592 | 0.23 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2067 |
0.32 |
| chr8_103802665_103802816 | 0.23 |
ENSG00000266799 |
. |
56793 |
0.11 |
| chr19_925725_925912 | 0.23 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
182 |
0.87 |
| chr7_153247150_153247301 | 0.23 |
DPP6 |
dipeptidyl-peptidase 6 |
336957 |
0.01 |
| chr3_47822404_47822881 | 0.23 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
765 |
0.62 |
| chr6_2970743_2970993 | 0.23 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
572 |
0.7 |
| chr16_75836248_75836399 | 0.23 |
ENSG00000199299 |
. |
58732 |
0.12 |
| chr13_99909205_99909490 | 0.23 |
GPR18 |
G protein-coupled receptor 18 |
1281 |
0.47 |
| chr4_123820303_123820454 | 0.22 |
NUDT6 |
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
14065 |
0.17 |
| chr5_88161245_88161396 | 0.22 |
MEF2C |
myocyte enhancer factor 2C |
12474 |
0.24 |
| chr22_43315092_43315243 | 0.22 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
26712 |
0.19 |
| chr1_93646822_93647050 | 0.22 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
651 |
0.44 |
| chr4_17615953_17616129 | 0.22 |
AC006160.5 |
|
153 |
0.58 |
| chr19_2562414_2562565 | 0.22 |
CTC-265F19.2 |
|
49371 |
0.09 |
| chr15_60882704_60882968 | 0.22 |
RORA |
RAR-related orphan receptor A |
1904 |
0.39 |
| chr10_677138_677289 | 0.22 |
RP11-809C18.3 |
|
2635 |
0.23 |
| chr17_26697729_26697880 | 0.22 |
VTN |
vitronectin |
39 |
0.9 |
| chr11_71934379_71934736 | 0.22 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
188 |
0.89 |
| chr1_192776419_192777001 | 0.22 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
1461 |
0.52 |
| chr5_55290006_55290408 | 0.22 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
585 |
0.51 |
| chr2_198064152_198064348 | 0.22 |
ANKRD44 |
ankyrin repeat domain 44 |
1488 |
0.35 |
| chr1_197167037_197167371 | 0.22 |
ZBTB41 |
zinc finger and BTB domain containing 41 |
2468 |
0.29 |
| chr15_60881660_60881911 | 0.22 |
RORA |
RAR-related orphan receptor A |
2955 |
0.3 |
| chr16_18957307_18957806 | 0.22 |
ENSG00000265515 |
. |
15952 |
0.13 |
| chr14_50160138_50160409 | 0.22 |
KLHDC1 |
kelch domain containing 1 |
426 |
0.71 |
| chr2_220113848_220114166 | 0.22 |
STK16 |
serine/threonine kinase 16 |
3389 |
0.09 |
| chr1_114413652_114413958 | 0.21 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
515 |
0.69 |
| chr18_55298168_55298387 | 0.21 |
RP11-35G9.3 |
|
695 |
0.64 |
| chr6_152505938_152506206 | 0.21 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
16573 |
0.28 |
| chr5_67577155_67577306 | 0.21 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1095 |
0.66 |
| chr1_19717605_19717920 | 0.21 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
5471 |
0.18 |
| chr6_135545194_135545568 | 0.21 |
ENSG00000207689 |
. |
14917 |
0.16 |
| chr12_69007685_69008546 | 0.21 |
RAP1B |
RAP1B, member of RAS oncogene family |
3310 |
0.27 |
| chr20_31099035_31099434 | 0.21 |
C20orf112 |
chromosome 20 open reading frame 112 |
24 |
0.98 |
| chr7_150264558_150265605 | 0.21 |
GIMAP4 |
GTPase, IMAP family member 4 |
557 |
0.78 |
| chr5_159898654_159898805 | 0.21 |
ENSG00000265237 |
. |
2680 |
0.24 |
| chr14_57283183_57283334 | 0.21 |
OTX2-AS1 |
OTX2 antisense RNA 1 (head to head) |
3249 |
0.2 |
| chr12_10328213_10328364 | 0.21 |
TMEM52B |
transmembrane protein 52B |
3324 |
0.16 |
| chr14_102278905_102279056 | 0.21 |
CTD-2017C7.2 |
|
2322 |
0.22 |
| chr7_21472009_21472160 | 0.21 |
SP4 |
Sp4 transcription factor |
4423 |
0.22 |
| chr12_93834472_93834763 | 0.21 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
415 |
0.81 |
| chr13_46755018_46755169 | 0.21 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1366 |
0.39 |
| chr14_22994017_22994177 | 0.20 |
TRAJ15 |
T cell receptor alpha joining 15 |
4483 |
0.13 |
| chr1_84610455_84610717 | 0.20 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
632 |
0.82 |
| chr10_53873012_53873163 | 0.20 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
66550 |
0.14 |
| chr12_53781982_53782414 | 0.20 |
SP1 |
Sp1 transcription factor |
7112 |
0.13 |
| chr3_183947999_183948171 | 0.20 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
132 |
0.92 |
| chr5_143571495_143571743 | 0.20 |
KCTD16 |
potassium channel tetramerization domain containing 16 |
13221 |
0.22 |
| chr8_135697544_135697755 | 0.20 |
ZFAT |
zinc finger and AT hook domain containing |
11148 |
0.28 |
| chr3_95425254_95425405 | 0.20 |
ENSG00000221477 |
. |
460417 |
0.01 |
| chr6_129828990_129829141 | 0.20 |
RP1-69D17.3 |
|
26510 |
0.24 |
| chr14_61809095_61809246 | 0.20 |
PRKCH |
protein kinase C, eta |
1790 |
0.38 |
| chr5_81074467_81075538 | 0.20 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr5_56794860_56795362 | 0.20 |
ACTBL2 |
actin, beta-like 2 |
16475 |
0.22 |
| chr15_101780403_101780609 | 0.20 |
CHSY1 |
chondroitin sulfate synthase 1 |
11631 |
0.18 |
| chr13_92500241_92500392 | 0.20 |
ENSG00000252508 |
. |
211781 |
0.03 |
| chr12_31957451_31957616 | 0.20 |
ENSG00000252421 |
. |
1509 |
0.36 |
| chr20_48381612_48381763 | 0.20 |
ENSG00000252540 |
. |
29661 |
0.13 |
| chr6_149663878_149664029 | 0.20 |
TAB2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
21765 |
0.18 |
| chrX_78417981_78418132 | 0.20 |
GPR174 |
G protein-coupled receptor 174 |
8413 |
0.33 |
| chr13_30419313_30419464 | 0.20 |
UBL3 |
ubiquitin-like 3 |
5433 |
0.32 |
| chrX_26102260_26102411 | 0.20 |
MAGEB18 |
melanoma antigen family B, 18 |
54125 |
0.17 |
| chr2_32390112_32390263 | 0.20 |
SLC30A6 |
solute carrier family 30 (zinc transporter), member 6 |
746 |
0.69 |
| chr14_102276711_102277223 | 0.19 |
CTD-2017C7.2 |
|
309 |
0.73 |
| chr1_38858671_38858822 | 0.19 |
ENSG00000200796 |
. |
3585 |
0.36 |
| chr3_63964903_63965338 | 0.19 |
ATXN7 |
ataxin 7 |
11700 |
0.14 |
| chr5_20958214_20958365 | 0.19 |
CDH18 |
cadherin 18, type 2 |
382307 |
0.01 |
| chr7_55842183_55842464 | 0.19 |
ENSG00000206729 |
. |
14738 |
0.2 |
| chr1_86315709_86315987 | 0.19 |
ENSG00000201620 |
. |
33633 |
0.23 |
| chr14_22342431_22342583 | 0.19 |
ENSG00000222776 |
. |
93722 |
0.08 |
| chr5_102295026_102295177 | 0.19 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
9820 |
0.31 |
| chr5_73932696_73932847 | 0.19 |
HEXB |
hexosaminidase B (beta polypeptide) |
3077 |
0.26 |
| chr6_111801598_111801749 | 0.19 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
2741 |
0.26 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 | 1.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.3 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.4 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0042510 | regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.2 | GO:2000351 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.3 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0090201 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.4 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.0 | GO:0002902 | regulation of B cell apoptotic process(GO:0002902) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043370) regulation of CD4-positive, alpha-beta T cell activation(GO:2000514) |
| 0.0 | 0.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |