Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
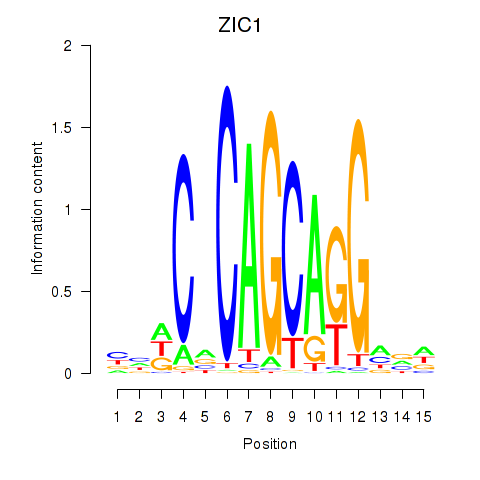
Results for ZIC1
Z-value: 1.28
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.5 | Zic family member 1 |
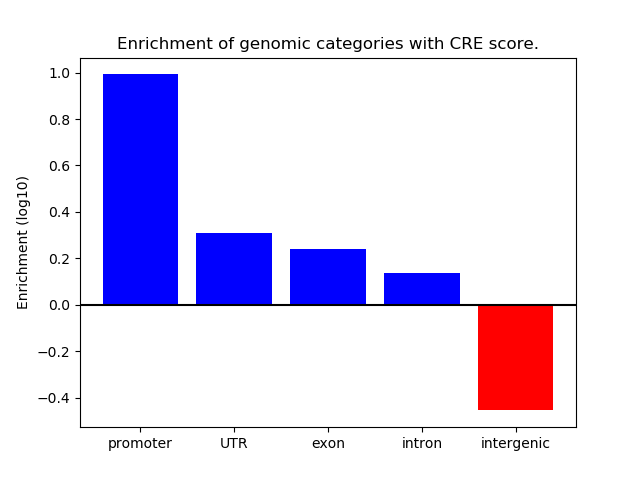
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_147131973_147132124 | ZIC1 | 4877 | 0.208815 | 0.84 | 4.2e-03 | Click! |
| chr3_147126089_147126240 | ZIC1 | 1007 | 0.450791 | 0.83 | 5.3e-03 | Click! |
| chr3_147142704_147142855 | ZIC1 | 15608 | 0.186611 | 0.82 | 6.9e-03 | Click! |
| chr3_147115262_147115580 | ZIC1 | 3739 | 0.210134 | 0.81 | 8.5e-03 | Click! |
| chr3_147111194_147111421 | ZIC1 | 375 | 0.850874 | 0.80 | 9.0e-03 | Click! |
Activity of the ZIC1 motif across conditions
Conditions sorted by the z-value of the ZIC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
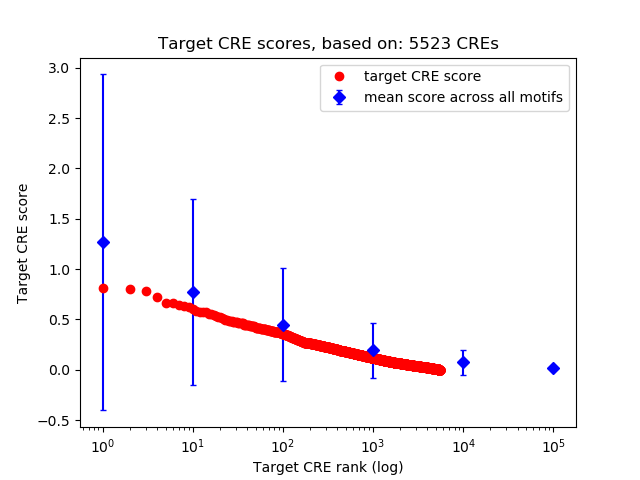
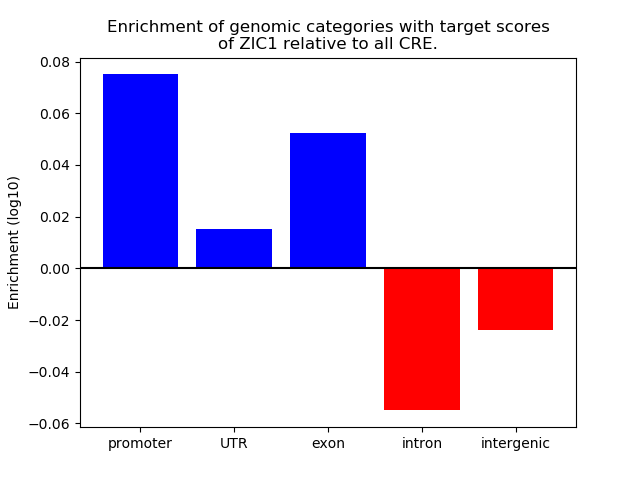
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_108520152_108520684 | 0.81 |
FAM155A |
family with sequence similarity 155, member A |
1335 |
0.55 |
| chr17_75317825_75318000 | 0.80 |
SEPT9 |
septin 9 |
780 |
0.69 |
| chr10_110225810_110226152 | 0.79 |
ENSG00000222436 |
. |
475087 |
0.01 |
| chr2_158113918_158114875 | 0.72 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
286 |
0.93 |
| chr12_81102183_81102935 | 0.67 |
MYF6 |
myogenic factor 6 (herculin) |
1282 |
0.49 |
| chr8_126013201_126013598 | 0.66 |
SQLE |
squalene epoxidase |
2613 |
0.23 |
| chr6_123110376_123110857 | 0.64 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
145 |
0.96 |
| chr9_12776208_12776436 | 0.63 |
LURAP1L |
leucine rich adaptor protein 1-like |
1302 |
0.47 |
| chr2_142887906_142888379 | 0.62 |
LRP1B |
low density lipoprotein receptor-related protein 1B |
432 |
0.87 |
| chr2_27301899_27302732 | 0.61 |
EMILIN1 |
elastin microfibril interfacer 1 |
880 |
0.32 |
| chr9_8857014_8857547 | 0.59 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
496 |
0.84 |
| chr17_29887299_29887674 | 0.58 |
ENSG00000207614 |
. |
471 |
0.69 |
| chr4_113739356_113739869 | 0.57 |
ANK2 |
ankyrin 2, neuronal |
347 |
0.9 |
| chr6_48037118_48037375 | 0.57 |
PTCHD4 |
patched domain containing 4 |
821 |
0.78 |
| chr2_235862821_235863118 | 0.55 |
SH3BP4 |
SH3-domain binding protein 4 |
2249 |
0.48 |
| chr6_12292037_12292313 | 0.55 |
EDN1 |
endothelin 1 |
1579 |
0.54 |
| chrX_8699533_8700279 | 0.54 |
KAL1 |
Kallmann syndrome 1 sequence |
321 |
0.94 |
| chr15_101513524_101514094 | 0.54 |
LRRK1 |
leucine-rich repeat kinase 1 |
54259 |
0.12 |
| chr12_110993281_110993585 | 0.53 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
27692 |
0.12 |
| chr20_34471632_34471916 | 0.52 |
ENSG00000201221 |
. |
3807 |
0.18 |
| chrX_117250265_117250710 | 0.51 |
KLHL13 |
kelch-like family member 13 |
274 |
0.95 |
| chrX_151142056_151142353 | 0.50 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
939 |
0.49 |
| chr22_46454527_46455045 | 0.50 |
RP6-109B7.3 |
|
3991 |
0.12 |
| chr3_9989057_9989268 | 0.50 |
PRRT3-AS1 |
PRRT3 antisense RNA 1 |
74 |
0.93 |
| chr5_73729921_73730212 | 0.49 |
ENSG00000244326 |
. |
116255 |
0.06 |
| chr5_166406240_166406516 | 0.48 |
TENM2 |
teneurin transmembrane protein 2 |
305426 |
0.01 |
| chr3_87040086_87040946 | 0.48 |
VGLL3 |
vestigial like 3 (Drosophila) |
247 |
0.96 |
| chr8_127837007_127837644 | 0.48 |
ENSG00000212451 |
. |
153558 |
0.04 |
| chr7_127899527_127899678 | 0.48 |
LEP |
leptin |
18265 |
0.16 |
| chr4_106816123_106816518 | 0.48 |
NPNT |
nephronectin |
274 |
0.81 |
| chr19_51161641_51162497 | 0.47 |
C19orf81 |
chromosome 19 open reading frame 81 |
9024 |
0.12 |
| chr9_130341109_130341430 | 0.47 |
FAM129B |
family with sequence similarity 129, member B |
1 |
0.98 |
| chr2_149894818_149895436 | 0.47 |
LYPD6B |
LY6/PLAUR domain containing 6B |
105 |
0.98 |
| chr14_71022448_71023169 | 0.47 |
ADAM20 |
ADAM metallopeptidase domain 20 |
21076 |
0.17 |
| chr4_175750179_175750687 | 0.46 |
GLRA3 |
glycine receptor, alpha 3 |
32 |
0.99 |
| chr20_9049976_9050220 | 0.46 |
PLCB4 |
phospholipase C, beta 4 |
350 |
0.89 |
| chr10_34815325_34815760 | 0.45 |
PARD3 |
par-3 family cell polarity regulator |
99883 |
0.09 |
| chr6_166075279_166075470 | 0.45 |
PDE10A |
phosphodiesterase 10A |
183 |
0.97 |
| chr1_16472641_16472879 | 0.45 |
RP11-276H7.2 |
|
8946 |
0.12 |
| chr3_8809480_8810320 | 0.44 |
OXTR |
oxytocin receptor |
463 |
0.76 |
| chr13_30157377_30157646 | 0.44 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
3418 |
0.36 |
| chr9_23821130_23821679 | 0.44 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
74 |
0.99 |
| chr17_28667391_28667637 | 0.44 |
ENSG00000207132 |
. |
4108 |
0.15 |
| chr10_6443642_6443853 | 0.44 |
DKFZP667F0711 |
|
51469 |
0.16 |
| chr19_14605464_14605615 | 0.44 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
1405 |
0.26 |
| chr2_10526122_10526400 | 0.44 |
HPCAL1 |
hippocalcin-like 1 |
33886 |
0.15 |
| chr14_89769991_89770787 | 0.44 |
RP11-356K23.1 |
|
46240 |
0.14 |
| chr7_116660327_116660575 | 0.43 |
ST7 |
suppression of tumorigenicity 7 |
120 |
0.96 |
| chr12_56075239_56075869 | 0.42 |
METTL7B |
methyltransferase like 7B |
224 |
0.86 |
| chr5_92908415_92908864 | 0.42 |
ENSG00000237187 |
. |
1590 |
0.42 |
| chr1_1283171_1284080 | 0.42 |
DVL1 |
dishevelled segment polarity protein 1 |
867 |
0.32 |
| chr8_41165761_41166040 | 0.42 |
SFRP1 |
secreted frizzled-related protein 1 |
1116 |
0.48 |
| chr4_138974934_138975159 | 0.42 |
ENSG00000250033 |
. |
3572 |
0.39 |
| chr6_10383738_10384217 | 0.42 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
20850 |
0.18 |
| chr12_54676753_54676980 | 0.42 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
884 |
0.34 |
| chr1_33367226_33367525 | 0.41 |
TMEM54 |
transmembrane protein 54 |
385 |
0.8 |
| chr9_102591075_102591263 | 0.41 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
2160 |
0.4 |
| chr11_77733968_77734525 | 0.41 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
94 |
0.72 |
| chr6_4369736_4370097 | 0.41 |
ENSG00000201185 |
. |
58281 |
0.15 |
| chr3_123476244_123476543 | 0.40 |
MYLK |
myosin light chain kinase |
36295 |
0.15 |
| chr1_170633420_170634117 | 0.40 |
PRRX1 |
paired related homeobox 1 |
690 |
0.8 |
| chr2_153193063_153193456 | 0.40 |
FMNL2 |
formin-like 2 |
1508 |
0.52 |
| chr20_47299051_47299422 | 0.40 |
ENSG00000251876 |
. |
56749 |
0.16 |
| chr6_76234416_76234599 | 0.40 |
FILIP1 |
filamin A interacting protein 1 |
31053 |
0.15 |
| chr7_93550787_93551002 | 0.40 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
117 |
0.96 |
| chr1_109490979_109491171 | 0.40 |
CLCC1 |
chloride channel CLIC-like 1 |
1985 |
0.29 |
| chr1_153581254_153581448 | 0.40 |
S100A16 |
S100 calcium binding protein A16 |
462 |
0.62 |
| chr9_132096752_132097176 | 0.40 |
ENSG00000242281 |
. |
35776 |
0.16 |
| chr11_122028989_122029168 | 0.40 |
ENSG00000207994 |
. |
6062 |
0.18 |
| chr8_94859053_94859497 | 0.40 |
TMEM67 |
transmembrane protein 67 |
61778 |
0.09 |
| chr11_61685093_61685479 | 0.39 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
289 |
0.85 |
| chr2_176931780_176932252 | 0.39 |
EVX2 |
even-skipped homeobox 2 |
16625 |
0.1 |
| chr1_201430589_201431038 | 0.39 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
7499 |
0.16 |
| chr5_33891538_33892159 | 0.39 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
198 |
0.94 |
| chr2_216296824_216297216 | 0.38 |
FN1 |
fibronectin 1 |
3770 |
0.26 |
| chr11_122046887_122047202 | 0.38 |
ENSG00000207994 |
. |
24028 |
0.16 |
| chr18_53087556_53087938 | 0.38 |
TCF4 |
transcription factor 4 |
1839 |
0.39 |
| chr1_13910795_13911459 | 0.38 |
PDPN |
podoplanin |
362 |
0.87 |
| chr11_131416298_131416516 | 0.38 |
AP003025.2 |
|
5646 |
0.24 |
| chr2_85632386_85633119 | 0.38 |
CAPG |
capping protein (actin filament), gelsolin-like |
3186 |
0.11 |
| chr11_2949356_2950534 | 0.38 |
PHLDA2 |
pleckstrin homology-like domain, family A, member 2 |
740 |
0.54 |
| chr4_38121545_38121863 | 0.38 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
74212 |
0.12 |
| chr1_60031328_60031726 | 0.38 |
FGGY |
FGGY carbohydrate kinase domain containing |
11937 |
0.3 |
| chr19_52222495_52223170 | 0.38 |
HAS1 |
hyaluronan synthase 1 |
110 |
0.94 |
| chr10_25155972_25156509 | 0.38 |
ENSG00000240294 |
. |
41501 |
0.18 |
| chr2_1711742_1712354 | 0.38 |
PXDN |
peroxidasin homolog (Drosophila) |
15706 |
0.26 |
| chr11_8832007_8832297 | 0.37 |
ST5 |
suppression of tumorigenicity 5 |
44 |
0.97 |
| chr9_94710782_94711296 | 0.37 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
123 |
0.98 |
| chr2_63893254_63893651 | 0.37 |
ENSG00000221085 |
. |
29217 |
0.21 |
| chr7_83278228_83278460 | 0.37 |
SEMA3E |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
18 |
0.99 |
| chr1_65774988_65775539 | 0.37 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
35 |
0.98 |
| chr2_196978205_196978455 | 0.37 |
RP11-347P5.1 |
|
37657 |
0.15 |
| chr7_117512754_117513170 | 0.37 |
CTTNBP2 |
cortactin binding protein 2 |
599 |
0.85 |
| chr3_134088468_134088746 | 0.36 |
AMOTL2 |
angiomotin like 2 |
2147 |
0.35 |
| chr20_8112947_8113200 | 0.36 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
199 |
0.97 |
| chr20_10651235_10651457 | 0.36 |
RP11-103J8.1 |
|
2230 |
0.35 |
| chr11_65259449_65259600 | 0.36 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
33024 |
0.08 |
| chr14_100204021_100204271 | 0.36 |
EML1 |
echinoderm microtubule associated protein like 1 |
60 |
0.98 |
| chr7_90084746_90084920 | 0.36 |
CDK14 |
cyclin-dependent kinase 14 |
10905 |
0.23 |
| chr3_45241366_45241603 | 0.36 |
TMEM158 |
transmembrane protein 158 (gene/pseudogene) |
26286 |
0.18 |
| chr19_13136060_13136211 | 0.36 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
324 |
0.8 |
| chr8_495850_496251 | 0.36 |
TDRP |
testis development related protein |
269 |
0.95 |
| chrX_153595297_153595831 | 0.36 |
FLNA |
filamin A, alpha |
4087 |
0.1 |
| chr7_89841494_89841721 | 0.35 |
STEAP2 |
STEAP family member 2, metalloreductase |
362 |
0.65 |
| chr14_85707273_85707789 | 0.35 |
ENSG00000252529 |
. |
30745 |
0.26 |
| chr1_112933403_112933636 | 0.35 |
CTTNBP2NL |
CTTNBP2 N-terminal like |
5284 |
0.23 |
| chr1_44300309_44300553 | 0.35 |
ST3GAL3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
64829 |
0.09 |
| chr6_38230060_38230313 | 0.35 |
ENSG00000200706 |
. |
55136 |
0.16 |
| chr12_77625749_77626115 | 0.35 |
ENSG00000238769 |
. |
69127 |
0.14 |
| chr2_181559754_181559963 | 0.34 |
ENSG00000264976 |
. |
77925 |
0.12 |
| chr9_35341457_35341792 | 0.34 |
AL160274.1 |
HCG17281; PRO0038; Uncharacterized protein |
19638 |
0.2 |
| chr3_64670816_64671530 | 0.34 |
ADAMTS9-AS2 |
ADAMTS9 antisense RNA 2 |
369 |
0.87 |
| chr6_27205685_27206711 | 0.34 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr4_74735225_74735723 | 0.34 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
353 |
0.85 |
| chr5_176899210_176899576 | 0.34 |
DBN1 |
drebrin 1 |
478 |
0.65 |
| chr2_79739509_79740065 | 0.34 |
CTNNA2 |
catenin (cadherin-associated protein), alpha 2 |
339 |
0.91 |
| chr19_48016527_48016998 | 0.34 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
703 |
0.59 |
| chr10_52834229_52834739 | 0.33 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
550 |
0.81 |
| chr17_74536772_74536989 | 0.33 |
PRCD |
progressive rod-cone degeneration |
708 |
0.43 |
| chr5_2752991_2753357 | 0.33 |
C5orf38 |
chromosome 5 open reading frame 38 |
795 |
0.6 |
| chr22_30601263_30601429 | 0.33 |
RP3-438O4.4 |
|
1752 |
0.27 |
| chr6_130450991_130451225 | 0.33 |
RP11-73O6.3 |
|
8233 |
0.25 |
| chr3_23713475_23713674 | 0.33 |
ENSG00000212269 |
. |
30491 |
0.19 |
| chr1_33207592_33208218 | 0.32 |
KIAA1522 |
KIAA1522 |
419 |
0.8 |
| chr7_36195246_36195841 | 0.32 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
2666 |
0.32 |
| chr1_167789688_167789943 | 0.32 |
RP1-313L4.3 |
|
171 |
0.96 |
| chr20_46412966_46413363 | 0.32 |
SULF2 |
sulfatase 2 |
1069 |
0.59 |
| chr12_113683537_113683756 | 0.32 |
TPCN1 |
two pore segment channel 1 |
1473 |
0.33 |
| chr18_421335_421729 | 0.32 |
RP11-720L2.2 |
|
2884 |
0.33 |
| chr2_218714763_218715120 | 0.32 |
TNS1 |
tensin 1 |
8058 |
0.27 |
| chr18_8948520_8948699 | 0.32 |
RP11-674N23.4 |
|
146955 |
0.04 |
| chr7_84815825_84816071 | 0.31 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
223 |
0.97 |
| chr14_96671190_96671644 | 0.31 |
RP11-404P21.8 |
Uncharacterized protein |
236 |
0.48 |
| chr3_42113480_42113939 | 0.31 |
TRAK1 |
trafficking protein, kinesin binding 1 |
18853 |
0.25 |
| chr11_57267285_57267491 | 0.31 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
1404 |
0.25 |
| chr22_31198410_31198813 | 0.31 |
OSBP2 |
oxysterol binding protein 2 |
462 |
0.84 |
| chr20_55800377_55800528 | 0.31 |
RP4-813D12.2 |
|
10524 |
0.2 |
| chr2_37870062_37870489 | 0.31 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
4526 |
0.29 |
| chr16_75284494_75285397 | 0.31 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
100 |
0.96 |
| chr16_65154751_65155029 | 0.30 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
943 |
0.75 |
| chr19_9473504_9474272 | 0.30 |
ZNF177 |
zinc finger protein 177 |
181 |
0.93 |
| chr5_1494740_1494891 | 0.30 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
29277 |
0.18 |
| chr10_77155014_77155181 | 0.30 |
ENSG00000237149 |
. |
6180 |
0.2 |
| chr5_136481655_136482067 | 0.30 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
167366 |
0.04 |
| chr9_130860823_130861938 | 0.30 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
544 |
0.57 |
| chr7_101268427_101268664 | 0.30 |
MYL10 |
myosin, light chain 10, regulatory |
4031 |
0.33 |
| chr17_71287404_71287786 | 0.30 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
19952 |
0.15 |
| chr12_5541333_5541550 | 0.30 |
NTF3 |
neurotrophin 3 |
162 |
0.98 |
| chr1_216773992_216774229 | 0.30 |
ESRRG |
estrogen-related receptor gamma |
122564 |
0.06 |
| chr8_66861458_66861787 | 0.29 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
72173 |
0.12 |
| chr6_47666429_47666580 | 0.29 |
GPR115 |
G protein-coupled receptor 115 |
215 |
0.94 |
| chrX_17395960_17396111 | 0.29 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
2492 |
0.32 |
| chr4_184797459_184798247 | 0.29 |
STOX2 |
storkhead box 2 |
28656 |
0.22 |
| chr6_143605233_143605462 | 0.29 |
AL031320.1 |
|
154228 |
0.03 |
| chr2_205410516_205411018 | 0.29 |
PARD3B |
par-3 family cell polarity regulator beta |
44 |
0.99 |
| chr2_133015015_133015166 | 0.28 |
ENSG00000221288 |
. |
437 |
0.85 |
| chr10_112159035_112159217 | 0.28 |
ENSG00000221359 |
. |
34873 |
0.17 |
| chr2_41837870_41838465 | 0.28 |
ENSG00000221372 |
. |
14641 |
0.29 |
| chr11_74022029_74022645 | 0.28 |
P4HA3 |
prolyl 4-hydroxylase, alpha polypeptide III |
348 |
0.87 |
| chr11_5618695_5618968 | 0.28 |
TRIM6 |
tripartite motif containing 6 |
795 |
0.34 |
| chr3_38150153_38150438 | 0.28 |
ENSG00000201965 |
. |
16590 |
0.14 |
| chr12_53443889_53444561 | 0.28 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
255 |
0.86 |
| chr1_30556615_30556878 | 0.28 |
ENSG00000222787 |
. |
198997 |
0.03 |
| chr1_59229210_59229361 | 0.28 |
ENSG00000264081 |
. |
1322 |
0.47 |
| chr19_16272076_16272227 | 0.28 |
CIB3 |
calcium and integrin binding family member 3 |
12135 |
0.11 |
| chr5_2752285_2752739 | 0.28 |
C5orf38 |
chromosome 5 open reading frame 38 |
133 |
0.91 |
| chr2_242499136_242499442 | 0.27 |
BOK-AS1 |
BOK antisense RNA 1 |
897 |
0.38 |
| chr4_100326128_100326285 | 0.27 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
25242 |
0.19 |
| chr5_36608256_36608472 | 0.27 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
67 |
0.99 |
| chr7_117468363_117468588 | 0.27 |
CTTNBP2 |
cortactin binding protein 2 |
36641 |
0.22 |
| chr4_23890798_23891066 | 0.27 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
726 |
0.8 |
| chr13_77457508_77457742 | 0.27 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
2900 |
0.26 |
| chr1_6795750_6796041 | 0.27 |
ENSG00000239166 |
. |
16884 |
0.16 |
| chr17_41327724_41327928 | 0.27 |
NBR1 |
neighbor of BRCA1 gene 1 |
4566 |
0.16 |
| chr3_141378576_141378773 | 0.27 |
RNF7 |
ring finger protein 7 |
78372 |
0.09 |
| chr14_77606808_77607442 | 0.27 |
ZDHHC22 |
zinc finger, DHHC-type containing 22 |
38 |
0.57 |
| chr18_47090550_47090839 | 0.27 |
LIPG |
lipase, endothelial |
1979 |
0.29 |
| chr8_141129048_141129349 | 0.27 |
C8orf17 |
chromosome 8 open reading frame 17 |
185782 |
0.03 |
| chr15_44450903_44451054 | 0.27 |
FRMD5 |
FERM domain containing 5 |
35666 |
0.19 |
| chr9_93834219_93834510 | 0.27 |
SYK |
spleen tyrosine kinase |
244594 |
0.02 |
| chr1_17560057_17560274 | 0.27 |
PADI1 |
peptidyl arginine deiminase, type I |
315 |
0.88 |
| chr3_185815817_185816010 | 0.27 |
ETV5 |
ets variant 5 |
10420 |
0.22 |
| chr8_118993051_118993436 | 0.27 |
EXT1 |
exostosin glycosyltransferase 1 |
129410 |
0.06 |
| chr1_144708044_144708206 | 0.27 |
RP11-640M9.2 |
|
92994 |
0.07 |
| chr9_36163620_36163822 | 0.27 |
CCIN |
calicin |
5668 |
0.19 |
| chr11_131618597_131618988 | 0.27 |
NTM |
neurotrimin |
87904 |
0.1 |
| chr17_42636239_42636390 | 0.27 |
FZD2 |
frizzled family receptor 2 |
1389 |
0.42 |
| chr8_35234925_35235076 | 0.27 |
UNC5D |
unc-5 homolog D (C. elegans) |
141707 |
0.05 |
| chr10_128947024_128947230 | 0.27 |
FAM196A |
family with sequence similarity 196, member A |
28146 |
0.24 |
| chr12_93293392_93293882 | 0.27 |
EEA1 |
early endosome antigen 1 |
29470 |
0.2 |
| chr10_24738095_24738342 | 0.27 |
KIAA1217 |
KIAA1217 |
144 |
0.97 |
| chr17_70636816_70636967 | 0.27 |
ENSG00000200783 |
. |
23400 |
0.24 |
| chr12_105626746_105626963 | 0.27 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
374 |
0.89 |
| chr22_43547615_43548743 | 0.27 |
TSPO |
translocator protein (18kDa) |
242 |
0.9 |
| chr11_73099035_73099239 | 0.27 |
RP11-809N8.2 |
|
8344 |
0.15 |
| chr2_219596210_219596511 | 0.27 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
5937 |
0.15 |
| chr4_24796744_24797030 | 0.26 |
SOD3 |
superoxide dismutase 3, extracellular |
198 |
0.97 |
| chr19_55917680_55917831 | 0.26 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
1332 |
0.21 |
| chr11_7013739_7013936 | 0.26 |
ZNF214 |
zinc finger protein 214 |
27629 |
0.13 |
| chr1_152488256_152488562 | 0.26 |
CRCT1 |
cysteine-rich C-terminal 1 |
1431 |
0.33 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.4 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.4 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.5 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.2 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.1 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.3 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.1 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.3 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.3 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.3 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 0.4 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.5 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.4 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.1 | 0.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.1 | 0.2 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.2 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.6 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.3 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.2 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.3 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.2 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.8 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:1903306 | negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.2 | GO:0036473 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0032373 | positive regulation of sterol transport(GO:0032373) positive regulation of cholesterol transport(GO:0032376) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0046398 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of protein processing(GO:0010955) negative regulation of complement activation(GO:0045916) negative regulation of protein maturation(GO:1903318) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.0 | 0.9 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0061515 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.0 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.4 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.1 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0042754 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) negative regulation of behavior(GO:0048521) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0099504 | synaptic vesicle cycle(GO:0099504) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.0 | GO:0046851 | negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 0.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.1 | GO:0019430 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0035338 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0043584 | nose development(GO:0043584) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.6 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 4.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.3 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |