Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
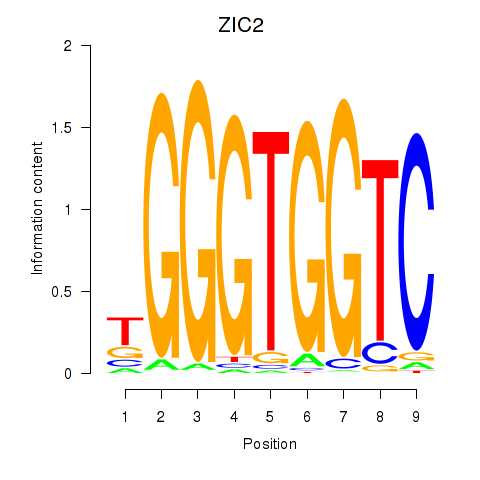
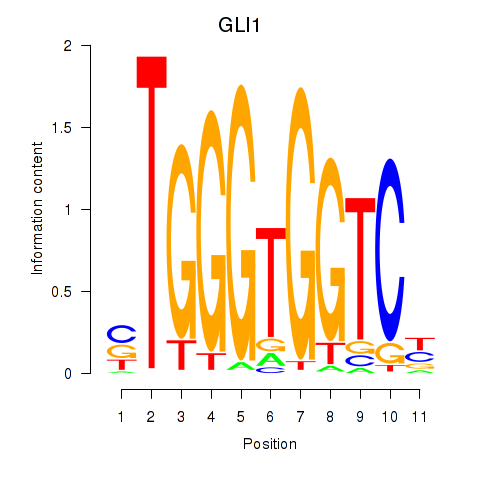
Results for ZIC2_GLI1
Z-value: 0.70
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
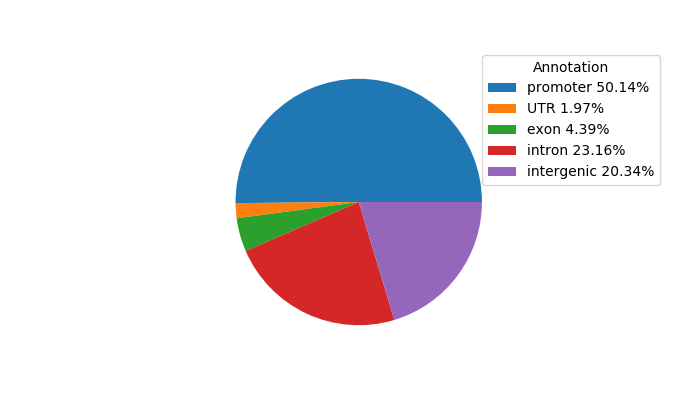
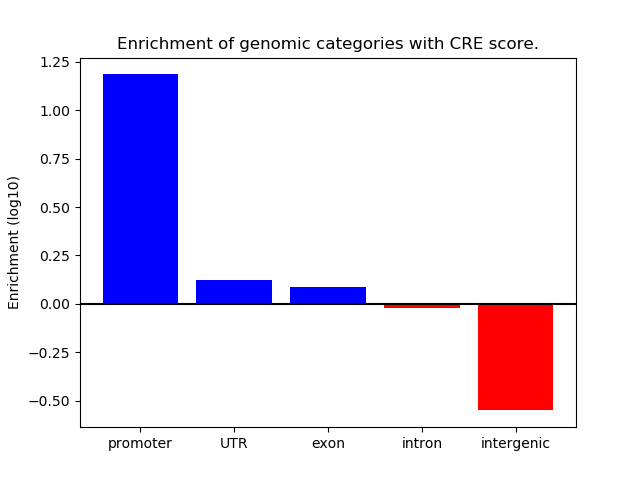
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_57854051_57854202 | GLI1 | 148 | 0.894550 | 0.53 | 1.5e-01 | Click! |
| chr12_57853426_57853753 | GLI1 | 329 | 0.745522 | 0.47 | 2.0e-01 | Click! |
| chr12_57854714_57854865 | GLI1 | 515 | 0.580095 | -0.40 | 2.9e-01 | Click! |
| chr12_57855715_57856187 | GLI1 | 1524 | 0.195259 | 0.34 | 3.7e-01 | Click! |
| chr12_57854208_57854360 | GLI1 | 10 | 0.943164 | 0.22 | 5.7e-01 | Click! |
| chr13_100631982_100632133 | ZIC2 | 1969 | 0.317336 | 0.68 | 4.3e-02 | Click! |
| chr13_100638993_100639144 | ZIC2 | 5042 | 0.202545 | -0.58 | 1.1e-01 | Click! |
| chr13_100632387_100632792 | ZIC2 | 1437 | 0.405386 | 0.55 | 1.3e-01 | Click! |
| chr13_100636352_100636503 | ZIC2 | 2401 | 0.278366 | 0.50 | 1.7e-01 | Click! |
| chr13_100633089_100633291 | ZIC2 | 836 | 0.610060 | 0.49 | 1.8e-01 | Click! |
Activity of the ZIC2_GLI1 motif across conditions
Conditions sorted by the z-value of the ZIC2_GLI1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
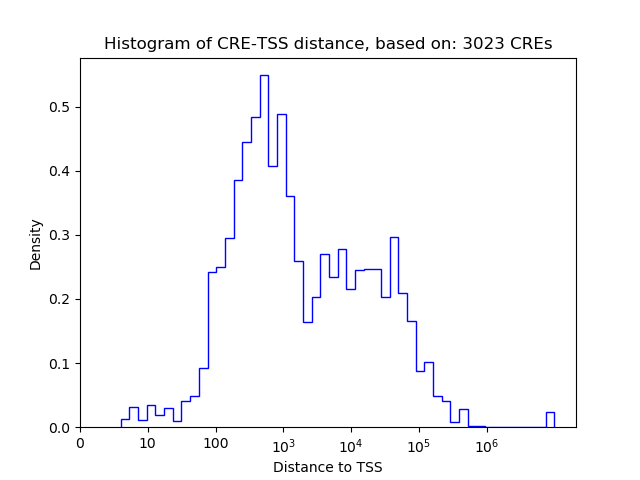
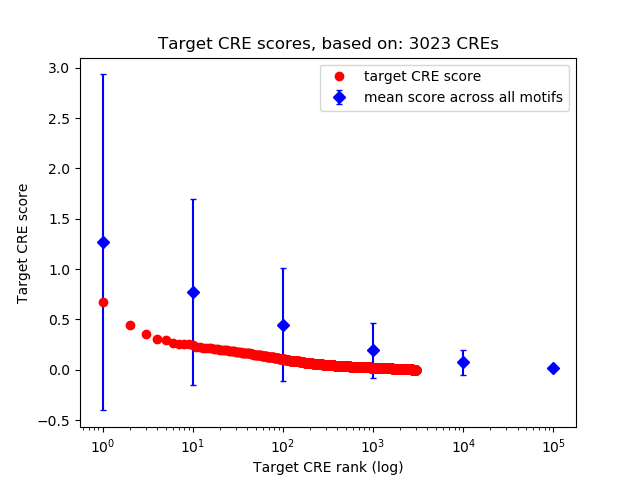
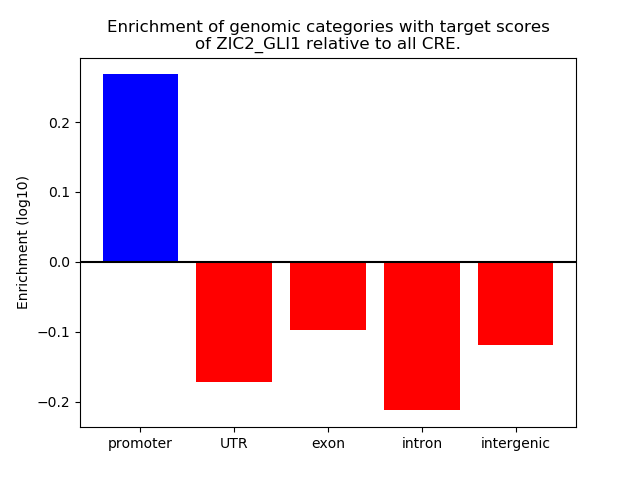
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_29836748_29836938 | 0.68 |
MVP |
major vault protein |
4090 |
0.07 |
| chr14_101294075_101294314 | 0.45 |
AL117190.2 |
|
1343 |
0.15 |
| chr9_132935666_132935932 | 0.36 |
NCS1 |
neuronal calcium sensor 1 |
942 |
0.6 |
| chr9_97906510_97906877 | 0.31 |
ENSG00000251884 |
. |
1870 |
0.3 |
| chr22_31502387_31502566 | 0.29 |
SELM |
Selenoprotein M |
1078 |
0.36 |
| chr10_5617288_5617517 | 0.27 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
49193 |
0.09 |
| chr7_120970338_120970562 | 0.26 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
1393 |
0.55 |
| chr11_27721206_27721474 | 0.26 |
BDNF |
brain-derived neurotrophic factor |
126 |
0.97 |
| chr1_214723918_214724069 | 0.26 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
573 |
0.84 |
| chr8_20121163_20121339 | 0.25 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
8448 |
0.2 |
| chr17_18062032_18062240 | 0.23 |
MYO15A |
myosin XVA |
484 |
0.71 |
| chrX_12809123_12809409 | 0.23 |
PRPS2 |
phosphoribosyl pyrophosphate synthetase 2 |
208 |
0.96 |
| chr15_71504267_71504460 | 0.22 |
RP11-673C5.2 |
|
22982 |
0.23 |
| chr3_187870883_187871050 | 0.21 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
106 |
0.93 |
| chr16_86613649_86613835 | 0.21 |
FOXL1 |
forkhead box L1 |
1627 |
0.34 |
| chr12_107297081_107297446 | 0.21 |
RP11-412D9.4 |
|
52012 |
0.12 |
| chr5_178956987_178957198 | 0.21 |
RUFY1 |
RUN and FYVE domain containing 1 |
20467 |
0.17 |
| chr19_13319766_13319917 | 0.21 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
48373 |
0.08 |
| chr5_60602887_60603064 | 0.21 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
25125 |
0.25 |
| chr6_170055214_170055550 | 0.20 |
WDR27 |
WD repeat domain 27 |
5406 |
0.19 |
| chr17_754296_754705 | 0.20 |
NXN |
nucleoredoxin |
12837 |
0.15 |
| chr3_147015992_147016163 | 0.19 |
RP11-649A16.1 |
|
72366 |
0.11 |
| chr6_35285702_35286007 | 0.19 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
8339 |
0.18 |
| chr1_45309015_45309207 | 0.19 |
PTCH2 |
patched 2 |
376 |
0.75 |
| chr1_214453711_214453918 | 0.19 |
SMYD2 |
SET and MYND domain containing 2 |
762 |
0.77 |
| chr2_69244184_69244393 | 0.19 |
ANTXR1 |
anthrax toxin receptor 1 |
3759 |
0.26 |
| chr4_72053226_72053895 | 0.18 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
516 |
0.88 |
| chr18_42030332_42030483 | 0.18 |
SETBP1 |
SET binding protein 1 |
229731 |
0.02 |
| chr11_34938171_34938707 | 0.18 |
PDHX |
pyruvate dehydrogenase complex, component X |
251 |
0.64 |
| chrX_49126619_49127035 | 0.18 |
PPP1R3F |
protein phosphatase 1, regulatory subunit 3F |
257 |
0.83 |
| chr4_187901794_187901964 | 0.18 |
ENSG00000252382 |
. |
123269 |
0.06 |
| chr17_10100912_10101063 | 0.18 |
GAS7 |
growth arrest-specific 7 |
881 |
0.65 |
| chr8_142069585_142069736 | 0.18 |
PTK2 |
protein tyrosine kinase 2 |
57451 |
0.11 |
| chr5_150460766_150460936 | 0.18 |
TNIP1 |
TNFAIP3 interacting protein 1 |
146 |
0.96 |
| chr12_13197333_13197564 | 0.17 |
KIAA1467 |
KIAA1467 |
230 |
0.92 |
| chr14_102786393_102786880 | 0.17 |
ZNF839 |
zinc finger protein 839 |
540 |
0.71 |
| chrX_329574_329729 | 0.17 |
PPP2R3B |
protein phosphatase 2, regulatory subunit B'', beta |
5128 |
0.27 |
| chrY_279574_279727 | 0.17 |
NA |
NA |
> 106 |
NA |
| chr2_32502966_32503192 | 0.17 |
YIPF4 |
Yip1 domain family, member 4 |
100 |
0.97 |
| chr15_68573802_68573977 | 0.17 |
FEM1B |
fem-1 homolog b (C. elegans) |
1592 |
0.39 |
| chr1_172110530_172111247 | 0.17 |
ENSG00000207949 |
. |
2841 |
0.22 |
| chr5_146783513_146783669 | 0.16 |
CTB-108O6.2 |
|
2268 |
0.31 |
| chr11_95887560_95887798 | 0.16 |
ENSG00000266192 |
. |
186923 |
0.03 |
| chr3_11314161_11314473 | 0.16 |
ATG7 |
autophagy related 7 |
208 |
0.96 |
| chr19_47163250_47163584 | 0.16 |
DACT3-AS1 |
DACT3 antisense RNA 1 |
204 |
0.83 |
| chr3_10028091_10028485 | 0.15 |
EMC3 |
ER membrane protein complex subunit 3 |
78 |
0.92 |
| chr17_27072661_27072812 | 0.15 |
TRAF4 |
TNF receptor-associated factor 4 |
1025 |
0.2 |
| chr3_10067619_10067972 | 0.15 |
FANCD2 |
Fanconi anemia, complementation group D2 |
303 |
0.79 |
| chr3_50378734_50378978 | 0.15 |
ZMYND10-AS1 |
ZMYND10 antisense RNA 1 |
319 |
0.54 |
| chr19_1017181_1017478 | 0.15 |
TMEM259 |
transmembrane protein 259 |
93 |
0.91 |
| chr13_111061542_111061693 | 0.15 |
ENSG00000238629 |
. |
4935 |
0.26 |
| chr6_26016611_26016935 | 0.15 |
HIST1H1A |
histone cluster 1, H1a |
1267 |
0.21 |
| chr15_40697977_40698740 | 0.15 |
IVD |
isovaleryl-CoA dehydrogenase |
83 |
0.94 |
| chrX_47930156_47930426 | 0.15 |
ZNF630 |
zinc finger protein 630 |
192 |
0.92 |
| chr10_12111309_12111515 | 0.14 |
DHTKD1 |
dehydrogenase E1 and transketolase domain containing 1 |
389 |
0.84 |
| chr16_8891744_8892217 | 0.14 |
PMM2 |
phosphomannomutase 2 |
256 |
0.68 |
| chr7_4680990_4681313 | 0.14 |
FOXK1 |
forkhead box K1 |
2237 |
0.38 |
| chr1_3285886_3286037 | 0.14 |
PRDM16 |
PR domain containing 16 |
27094 |
0.21 |
| chr5_98109376_98109557 | 0.14 |
RGMB |
repulsive guidance molecule family member b |
127 |
0.72 |
| chr5_97550577_97550728 | 0.14 |
ENSG00000223053 |
. |
424283 |
0.01 |
| chr17_34115736_34115943 | 0.14 |
MMP28 |
matrix metallopeptidase 28 |
6872 |
0.1 |
| chr15_38117163_38117383 | 0.14 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
96867 |
0.09 |
| chr5_179248266_179248778 | 0.13 |
SQSTM1 |
sequestosome 1 |
601 |
0.58 |
| chr1_67142291_67142442 | 0.13 |
AL139147.1 |
Uncharacterized protein |
344 |
0.88 |
| chr10_51622913_51623296 | 0.13 |
TIMM23 |
translocase of inner mitochondrial membrane 23 homolog (yeast) |
99 |
0.97 |
| chr15_90357053_90357234 | 0.13 |
ANPEP |
alanyl (membrane) aminopeptidase |
951 |
0.46 |
| chrX_134166936_134167548 | 0.13 |
FAM127A |
family with sequence similarity 127, member A |
909 |
0.53 |
| chr1_20834735_20834945 | 0.13 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
186 |
0.95 |
| chr17_38225914_38226065 | 0.13 |
THRA |
thyroid hormone receptor, alpha |
833 |
0.48 |
| chr22_37699623_37699774 | 0.13 |
CYTH4 |
cytohesin 4 |
6311 |
0.18 |
| chr7_38350810_38351032 | 0.13 |
STARD3NL |
STARD3 N-terminal like |
132924 |
0.05 |
| chr7_101579659_101580126 | 0.13 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr20_45887719_45887887 | 0.13 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
39841 |
0.12 |
| chr8_102121369_102121647 | 0.13 |
ENSG00000202360 |
. |
28679 |
0.21 |
| chr14_24584036_24584230 | 0.13 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
63 |
0.51 |
| chr2_189748856_189749076 | 0.13 |
AC079613.1 |
|
44795 |
0.16 |
| chr10_51371455_51372104 | 0.12 |
TIMM23B |
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
343 |
0.58 |
| chr17_63096937_63097327 | 0.12 |
RGS9 |
regulator of G-protein signaling 9 |
36434 |
0.15 |
| chr10_75547124_75547275 | 0.12 |
ZSWIM8 |
zinc finger, SWIM-type containing 8 |
1779 |
0.16 |
| chr8_96146036_96146378 | 0.12 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
97 |
0.97 |
| chr1_55681610_55681784 | 0.12 |
USP24 |
ubiquitin specific peptidase 24 |
911 |
0.63 |
| chr7_130131272_130131861 | 0.12 |
MEST |
mesoderm specific transcript |
368 |
0.77 |
| chr19_44036829_44037014 | 0.12 |
ZNF575 |
zinc finger protein 575 |
195 |
0.88 |
| chr22_42679165_42679458 | 0.12 |
TCF20 |
transcription factor 20 (AR1) |
60311 |
0.1 |
| chr16_66440546_66441157 | 0.12 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
11493 |
0.17 |
| chr1_201425782_201426096 | 0.12 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
12373 |
0.15 |
| chr9_20340314_20340473 | 0.12 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
42074 |
0.15 |
| chr5_158326789_158326940 | 0.12 |
CTD-2363C16.1 |
|
83150 |
0.1 |
| chr9_98278987_98279233 | 0.12 |
PTCH1 |
patched 1 |
137 |
0.95 |
| chr4_110354244_110354664 | 0.11 |
SEC24B-AS1 |
SEC24B antisense RNA 1 |
466 |
0.46 |
| chr4_1863859_1864305 | 0.11 |
LETM1 |
leucine zipper-EF-hand containing transmembrane protein 1 |
6108 |
0.16 |
| chr10_81002854_81003026 | 0.11 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
63035 |
0.13 |
| chr20_36149696_36149913 | 0.11 |
NNAT |
neuronatin |
183 |
0.64 |
| chr2_114648081_114648567 | 0.11 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
149 |
0.97 |
| chr19_16435025_16435277 | 0.11 |
CTD-2562J15.6 |
|
174 |
0.78 |
| chr20_5485005_5485184 | 0.11 |
RP5-1022P6.5 |
|
29180 |
0.21 |
| chr11_65682969_65683236 | 0.11 |
C11orf68 |
chromosome 11 open reading frame 68 |
2729 |
0.12 |
| chr20_62257766_62257982 | 0.11 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
520 |
0.48 |
| chr17_39969662_39969906 | 0.10 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
601 |
0.45 |
| chr11_89867468_89867846 | 0.10 |
NAALAD2 |
N-acetylated alpha-linked acidic dipeptidase 2 |
40 |
0.98 |
| chr8_144922083_144922496 | 0.10 |
NRBP2 |
nuclear receptor binding protein 2 |
513 |
0.61 |
| chr17_61850012_61850913 | 0.10 |
CCDC47 |
coiled-coil domain containing 47 |
468 |
0.45 |
| chr7_26903418_26904290 | 0.10 |
SKAP2 |
src kinase associated phosphoprotein 2 |
508 |
0.86 |
| chr1_173794371_173794522 | 0.10 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
649 |
0.46 |
| chr12_32260246_32261058 | 0.10 |
RP11-843B15.2 |
|
190 |
0.75 |
| chr17_77966356_77966507 | 0.10 |
CTD-2529O21.1 |
|
209 |
0.88 |
| chr19_927045_927244 | 0.10 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
1144 |
0.25 |
| chr1_154531550_154531753 | 0.10 |
UBE2Q1 |
ubiquitin-conjugating enzyme E2Q family member 1 |
542 |
0.64 |
| chr2_26987219_26988126 | 0.10 |
SLC35F6 |
solute carrier family 35, member F6 |
515 |
0.78 |
| chr4_183481639_183481857 | 0.10 |
ENSG00000252343 |
. |
11936 |
0.26 |
| chr3_120149251_120149402 | 0.10 |
FSTL1 |
follistatin-like 1 |
20512 |
0.22 |
| chr16_82431120_82431271 | 0.10 |
ENSG00000264502 |
. |
50011 |
0.15 |
| chr12_26986439_26986924 | 0.10 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
550 |
0.83 |
| chr6_24910461_24910676 | 0.10 |
FAM65B |
family with sequence similarity 65, member B |
627 |
0.77 |
| chr14_72053260_72053532 | 0.10 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
398 |
0.92 |
| chr3_127872049_127872245 | 0.10 |
EEFSEC |
eukaryotic elongation factor, selenocysteine-tRNA-specific |
150 |
0.86 |
| chrX_47518304_47518499 | 0.09 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
115 |
0.54 |
| chr10_116020157_116020308 | 0.09 |
VWA2 |
von Willebrand factor A domain containing 2 |
21143 |
0.21 |
| chr4_56212171_56212371 | 0.09 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
5 |
0.98 |
| chr16_27325280_27325496 | 0.09 |
IL4R |
interleukin 4 receptor |
75 |
0.97 |
| chr8_17555063_17555276 | 0.09 |
MTUS1 |
microtubule associated tumor suppressor 1 |
10 |
0.97 |
| chr12_132688811_132688962 | 0.09 |
GALNT9 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
1451 |
0.44 |
| chr3_112358415_112358703 | 0.09 |
CCDC80 |
coiled-coil domain containing 80 |
1557 |
0.48 |
| chr16_75328031_75328182 | 0.09 |
ENSG00000252101 |
. |
7410 |
0.15 |
| chr3_49592272_49592564 | 0.09 |
BSN |
bassoon presynaptic cytomatrix protein |
496 |
0.78 |
| chr7_77326845_77327188 | 0.09 |
RSBN1L |
round spermatid basic protein 1-like |
548 |
0.83 |
| chr3_142956189_142956340 | 0.09 |
ENSG00000211483 |
. |
71774 |
0.1 |
| chr22_46473276_46473626 | 0.09 |
FLJ27365 |
hsa-mir-4763 |
2741 |
0.14 |
| chr20_62257208_62257699 | 0.09 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
941 |
0.32 |
| chr15_90356636_90356804 | 0.09 |
ANPEP |
alanyl (membrane) aminopeptidase |
1374 |
0.33 |
| chr1_23079970_23080247 | 0.09 |
ENSG00000216157 |
. |
4704 |
0.19 |
| chr2_46774565_46774716 | 0.09 |
RHOQ |
ras homolog family member Q |
4101 |
0.17 |
| chr6_85248911_85249062 | 0.09 |
RP11-132M7.3 |
|
150157 |
0.05 |
| chr13_114823633_114823784 | 0.09 |
RASA3 |
RAS p21 protein activator 3 |
19730 |
0.23 |
| chr16_85795783_85795934 | 0.09 |
C16orf74 |
chromosome 16 open reading frame 74 |
11123 |
0.1 |
| chr15_78171702_78171998 | 0.09 |
CSPG4P13 |
chondroitin sulfate proteoglycan 4 pseudogene 13 |
15176 |
0.15 |
| chr9_130854743_130854894 | 0.09 |
SLC25A25 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
1075 |
0.29 |
| chr3_72640881_72641032 | 0.09 |
ENSG00000199469 |
. |
15114 |
0.22 |
| chr8_47829477_47829948 | 0.09 |
ENSG00000252594 |
. |
87040 |
0.1 |
| chr19_980574_980725 | 0.09 |
WDR18 |
WD repeat domain 18 |
3682 |
0.1 |
| chr1_55352544_55352822 | 0.09 |
RP11-67L3.5 |
|
135 |
0.55 |
| chr1_72749537_72750410 | 0.09 |
NEGR1 |
neuronal growth regulator 1 |
1556 |
0.58 |
| chr11_46300240_46300518 | 0.09 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
1151 |
0.46 |
| chr3_71804156_71804307 | 0.09 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
307 |
0.82 |
| chr11_129764495_129764646 | 0.09 |
NFRKB |
nuclear factor related to kappaB binding protein |
234 |
0.95 |
| chr9_117149358_117150269 | 0.08 |
AKNA |
AT-hook transcription factor |
430 |
0.85 |
| chr7_130791022_130791700 | 0.08 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
428 |
0.85 |
| chr8_32406806_32407051 | 0.08 |
NRG1 |
neuregulin 1 |
683 |
0.82 |
| chr16_18494169_18494622 | 0.08 |
PKD1P5 |
polycystic kidney disease 1 (autosomal dominant) pseudogene 5 |
1402 |
0.22 |
| chr9_97818540_97818804 | 0.08 |
ENSG00000207563 |
. |
28818 |
0.12 |
| chr8_38964877_38965090 | 0.08 |
ADAM32 |
ADAM metallopeptidase domain 32 |
65 |
0.98 |
| chr8_22102673_22103420 | 0.08 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
298 |
0.63 |
| chr15_95847267_95847418 | 0.08 |
ENSG00000222076 |
. |
441691 |
0.01 |
| chr7_20370368_20370519 | 0.08 |
ITGB8 |
integrin, beta 8 |
118 |
0.93 |
| chr7_65670330_65671074 | 0.08 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
398 |
0.87 |
| chr16_56672198_56672419 | 0.08 |
MT1A |
metallothionein 1A |
270 |
0.75 |
| chr3_49449862_49450835 | 0.08 |
TCTA |
T-cell leukemia translocation altered |
709 |
0.34 |
| chr1_6674083_6674241 | 0.08 |
PHF13 |
PHD finger protein 13 |
417 |
0.47 |
| chr8_37246004_37246155 | 0.08 |
RP11-150O12.6 |
|
128460 |
0.05 |
| chr2_189844043_189844303 | 0.08 |
ENSG00000221502 |
. |
1355 |
0.48 |
| chr7_155436479_155436836 | 0.08 |
AC009403.2 |
Protein LOC100506302 |
438 |
0.51 |
| chr5_158878566_158878856 | 0.08 |
ENSG00000266432 |
. |
74235 |
0.11 |
| chr1_206879659_206879853 | 0.08 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
21374 |
0.13 |
| chr20_35171113_35171392 | 0.08 |
MYL9 |
myosin, light chain 9, regulatory |
1352 |
0.35 |
| chr2_20271939_20272144 | 0.08 |
RP11-644K8.1 |
|
20146 |
0.16 |
| chr7_134472278_134472429 | 0.08 |
CALD1 |
caldesmon 1 |
7924 |
0.3 |
| chr11_111411757_111412020 | 0.08 |
LAYN |
layilin |
170 |
0.92 |
| chr13_108022701_108022852 | 0.07 |
ENSG00000201847 |
. |
49465 |
0.19 |
| chr3_64430283_64430501 | 0.07 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
666 |
0.79 |
| chr17_55055562_55056181 | 0.07 |
SCPEP1 |
serine carboxypeptidase 1 |
376 |
0.79 |
| chr2_9983595_9984276 | 0.07 |
TAF1B |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa |
283 |
0.93 |
| chr17_43205760_43205911 | 0.07 |
PLCD3 |
phospholipase C, delta 3 |
4056 |
0.12 |
| chr17_6346848_6347134 | 0.07 |
FAM64A |
family with sequence similarity 64, member A |
744 |
0.65 |
| chr11_35676290_35676441 | 0.07 |
TRIM44 |
tripartite motif containing 44 |
7988 |
0.24 |
| chr3_147109765_147110149 | 0.07 |
ZIC1 |
Zic family member 1 |
1725 |
0.33 |
| chr20_42143535_42143824 | 0.07 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
513 |
0.75 |
| chr12_53441858_53442099 | 0.07 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
755 |
0.42 |
| chr12_121974248_121974441 | 0.07 |
KDM2B |
lysine (K)-specific demethylase 2B |
204 |
0.95 |
| chr8_67525864_67526623 | 0.07 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
714 |
0.66 |
| chr1_153584868_153585142 | 0.07 |
S100A16 |
S100 calcium binding protein A16 |
554 |
0.53 |
| chr9_123476196_123476418 | 0.07 |
MEGF9 |
multiple EGF-like-domains 9 |
305 |
0.93 |
| chr9_124131284_124131757 | 0.07 |
STOM |
stomatin |
963 |
0.5 |
| chr5_325316_325467 | 0.07 |
AHRR |
aryl-hydrocarbon receptor repressor |
2636 |
0.26 |
| chr11_69460305_69460629 | 0.07 |
CCND1 |
cyclin D1 |
4493 |
0.24 |
| chr20_18477327_18477868 | 0.07 |
RBBP9 |
retinoblastoma binding protein 9 |
290 |
0.85 |
| chr4_8595158_8595423 | 0.07 |
CPZ |
carboxypeptidase Z |
807 |
0.68 |
| chr10_52751128_52751393 | 0.07 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
142 |
0.97 |
| chr17_75318499_75318729 | 0.07 |
SEPT9 |
septin 9 |
78 |
0.98 |
| chr10_86056124_86056314 | 0.07 |
CCSER2 |
coiled-coil serine-rich protein 2 |
32123 |
0.16 |
| chr20_56884148_56884425 | 0.07 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
209 |
0.75 |
| chr10_180203_180821 | 0.07 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
88 |
0.98 |
| chr2_24270267_24270418 | 0.07 |
C2orf44 |
chromosome 2 open reading frame 44 |
111 |
0.92 |
| chr19_47735595_47736220 | 0.07 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
| chr9_91661673_91661920 | 0.07 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
4744 |
0.25 |
| chr16_83987560_83987743 | 0.07 |
OSGIN1 |
oxidative stress induced growth inhibitor 1 |
807 |
0.57 |
| chr6_21886939_21887090 | 0.07 |
ENSG00000222515 |
. |
198759 |
0.03 |
| chr19_5292236_5292538 | 0.07 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
411 |
0.89 |
| chr9_91429388_91429539 | 0.07 |
ENSG00000265873 |
. |
68643 |
0.13 |
| chr17_4710527_4711001 | 0.07 |
PLD2 |
phospholipase D2 |
93 |
0.93 |
| chr6_155635765_155636118 | 0.06 |
TFB1M |
transcription factor B1, mitochondrial |
323 |
0.92 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0071265 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0060073 | micturition(GO:0060073) |
| 0.0 | 0.0 | GO:0009111 | vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |