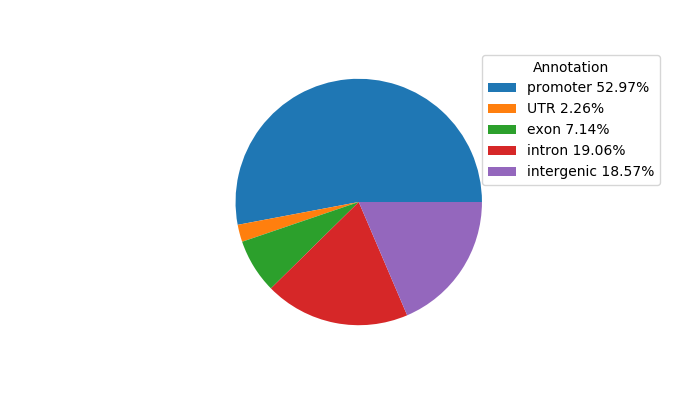
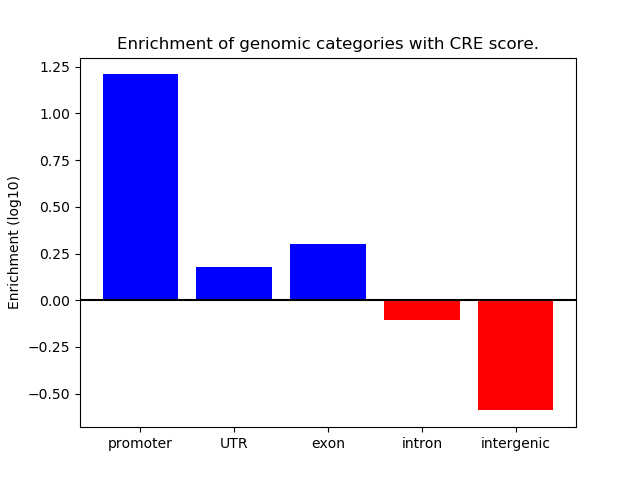
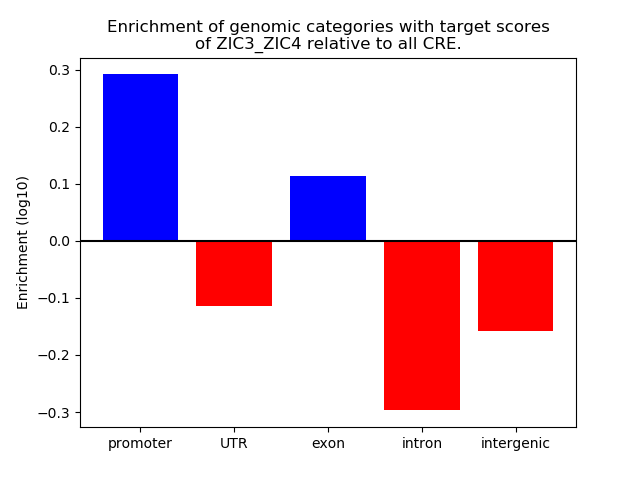
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ZIC3_ZIC4
Z-value: 0.79
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
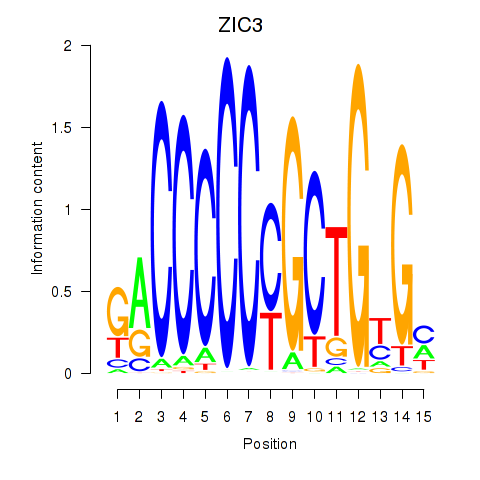
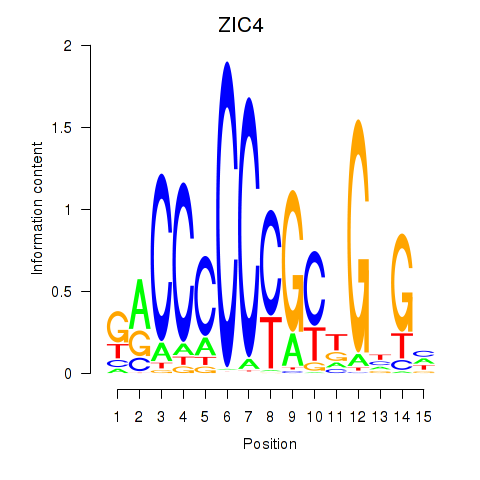
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
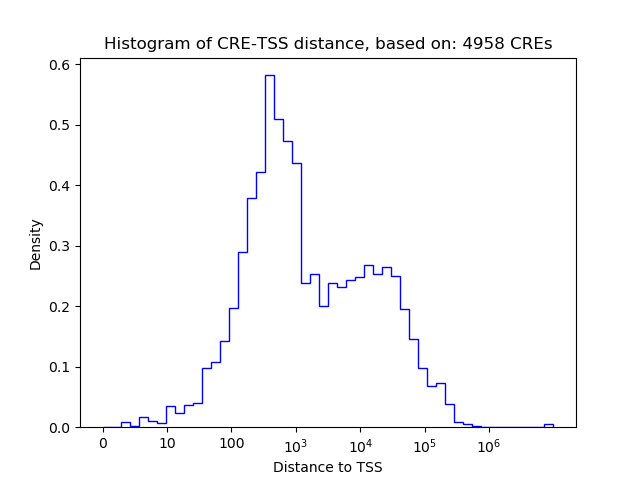
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_136650028_136650459 | ZIC3 | 1566 | 0.394980 | 0.53 | 1.4e-01 | Click! |
| chrX_136649219_136649370 | ZIC3 | 617 | 0.726656 | -0.41 | 2.8e-01 | Click! |
| chrX_136651041_136651192 | ZIC3 | 2439 | 0.293855 | -0.29 | 4.4e-01 | Click! |
| chrX_136647691_136647934 | ZIC3 | 489 | 0.730082 | 0.28 | 4.7e-01 | Click! |
| chrX_136650504_136650655 | ZIC3 | 1902 | 0.343807 | 0.14 | 7.1e-01 | Click! |
| chr3_147125315_147125495 | ZIC4 | 758 | 0.586247 | 0.43 | 2.5e-01 | Click! |
| chr3_147125608_147125766 | ZIC4 | 1040 | 0.434514 | 0.34 | 3.7e-01 | Click! |
| chr3_147125110_147125261 | ZIC4 | 538 | 0.729018 | 0.20 | 6.1e-01 | Click! |
Activity of the ZIC3_ZIC4 motif across conditions
Conditions sorted by the z-value of the ZIC3_ZIC4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
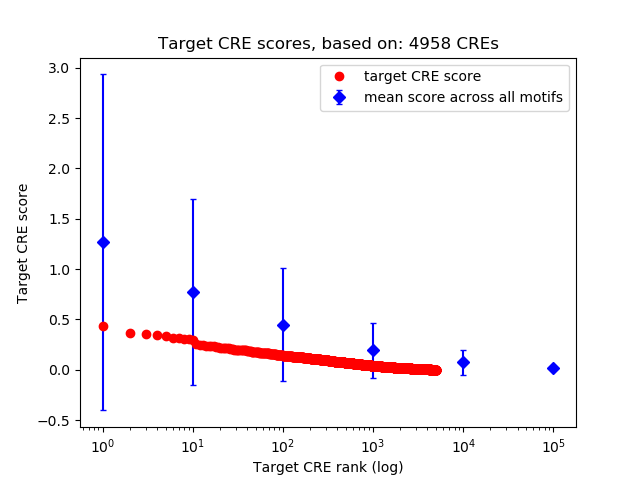
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_110872434_110873339 | 0.44 |
MALL |
mal, T-cell differentiation protein-like |
713 |
0.63 |
| chr2_236401822_236402641 | 0.37 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
502 |
0.83 |
| chr20_60941163_60941716 | 0.36 |
LAMA5 |
laminin, alpha 5 |
887 |
0.46 |
| chr1_201616947_201617125 | 0.34 |
NAV1 |
neuron navigator 1 |
341 |
0.86 |
| chr6_33387553_33387744 | 0.34 |
SYNGAP1 |
synaptic Ras GTPase activating protein 1 |
199 |
0.87 |
| chr4_184094744_184094905 | 0.32 |
ENSG00000252048 |
. |
3260 |
0.22 |
| chr13_24477780_24477964 | 0.31 |
C1QTNF9B |
C1q and tumor necrosis factor related protein 9B |
1078 |
0.48 |
| chr19_3688376_3688653 | 0.31 |
PIP5K1C |
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma |
11874 |
0.1 |
| chr17_38599985_38600404 | 0.31 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
481 |
0.74 |
| chr10_128250315_128250594 | 0.30 |
ENSG00000221717 |
. |
4821 |
0.29 |
| chr14_24837426_24838101 | 0.26 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
216 |
0.85 |
| chr9_112262221_112262626 | 0.25 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
1833 |
0.4 |
| chr20_30457693_30458808 | 0.24 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr15_89248242_89248620 | 0.24 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
66247 |
0.09 |
| chr8_143533593_143534231 | 0.23 |
BAI1 |
brain-specific angiogenesis inhibitor 1 |
3121 |
0.27 |
| chr20_25565265_25566139 | 0.23 |
NINL |
ninein-like |
451 |
0.81 |
| chr4_7972052_7972961 | 0.23 |
AFAP1 |
actin filament associated protein 1 |
30853 |
0.14 |
| chr8_102092961_102093560 | 0.23 |
ENSG00000202360 |
. |
56927 |
0.13 |
| chr22_44222171_44222754 | 0.23 |
RP3-388M5.9 |
|
14089 |
0.16 |
| chr2_23608861_23609561 | 0.22 |
KLHL29 |
kelch-like family member 29 |
1123 |
0.67 |
| chrX_73639854_73640301 | 0.22 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
1008 |
0.55 |
| chr1_156646561_156647039 | 0.22 |
NES |
nestin |
389 |
0.72 |
| chr17_48625105_48625289 | 0.21 |
SPATA20 |
spermatogenesis associated 20 |
677 |
0.5 |
| chr7_391737_391888 | 0.21 |
AC187652.1 |
Protein LOC100996433 |
88894 |
0.08 |
| chr17_38721330_38721617 | 0.21 |
CCR7 |
chemokine (C-C motif) receptor 7 |
238 |
0.92 |
| chr13_113777141_113777292 | 0.21 |
F10 |
coagulation factor X |
84 |
0.95 |
| chr8_22140544_22140695 | 0.21 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
7539 |
0.14 |
| chr19_3606102_3607092 | 0.20 |
TBXA2R |
thromboxane A2 receptor |
61 |
0.89 |
| chr1_1410134_1410538 | 0.20 |
ATAD3B |
ATPase family, AAA domain containing 3B |
3187 |
0.12 |
| chr6_151562009_151562901 | 0.20 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
946 |
0.51 |
| chr7_155598385_155598815 | 0.20 |
SHH |
sonic hedgehog |
3166 |
0.3 |
| chr19_10024160_10024434 | 0.20 |
OLFM2 |
olfactomedin 2 |
215 |
0.91 |
| chr10_110671781_110671954 | 0.20 |
ENSG00000222436 |
. |
29201 |
0.27 |
| chr16_66878332_66879083 | 0.20 |
CA7 |
carbonic anhydrase VII |
128 |
0.94 |
| chrX_30326616_30326849 | 0.20 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
127 |
0.96 |
| chr16_4364778_4365572 | 0.20 |
GLIS2 |
GLIS family zinc finger 2 |
413 |
0.78 |
| chr5_126625896_126626411 | 0.19 |
MEGF10 |
multiple EGF-like-domains 10 |
370 |
0.91 |
| chr11_66233628_66233779 | 0.19 |
PELI3 |
pellino E3 ubiquitin protein ligase family member 3 |
513 |
0.59 |
| chr8_41510934_41511242 | 0.19 |
NKX6-3 |
NK6 homeobox 3 |
2233 |
0.21 |
| chr19_42497759_42498630 | 0.19 |
ATP1A3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
37 |
0.97 |
| chr19_49561050_49561269 | 0.19 |
CGB7 |
chorionic gonadotropin, beta polypeptide 7 |
151 |
0.85 |
| chr3_183948174_183948496 | 0.19 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
118 |
0.93 |
| chr9_1051774_1052480 | 0.19 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
513 |
0.84 |
| chr7_117512754_117513170 | 0.18 |
CTTNBP2 |
cortactin binding protein 2 |
599 |
0.85 |
| chr1_33207034_33207258 | 0.18 |
KIAA1522 |
KIAA1522 |
340 |
0.85 |
| chr12_124871047_124871198 | 0.18 |
NCOR2 |
nuclear receptor corepressor 2 |
2248 |
0.42 |
| chr1_176748344_176748495 | 0.18 |
ENSG00000202609 |
. |
250162 |
0.02 |
| chr17_59481988_59482187 | 0.18 |
RP11-332H18.5 |
|
4483 |
0.14 |
| chr17_20491916_20492779 | 0.18 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr19_8675123_8675468 | 0.18 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
290 |
0.88 |
| chr19_14585851_14586099 | 0.18 |
PTGER1 |
prostaglandin E receptor 1 (subtype EP1), 42kDa |
199 |
0.89 |
| chr7_150671802_150672427 | 0.18 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
2900 |
0.18 |
| chr13_24007256_24007423 | 0.17 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
502 |
0.88 |
| chrX_53349448_53350467 | 0.17 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
565 |
0.73 |
| chr15_22893749_22894042 | 0.17 |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
1179 |
0.51 |
| chr4_2283428_2283729 | 0.17 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
5864 |
0.13 |
| chr10_90911880_90912031 | 0.17 |
CH25H |
cholesterol 25-hydroxylase |
55116 |
0.1 |
| chr3_71149968_71150208 | 0.17 |
FOXP1 |
forkhead box P1 |
29656 |
0.25 |
| chr10_28287400_28288216 | 0.17 |
ARMC4 |
armadillo repeat containing 4 |
169 |
0.97 |
| chr2_23607446_23607927 | 0.17 |
KLHL29 |
kelch-like family member 29 |
402 |
0.92 |
| chr7_94287847_94287998 | 0.17 |
PEG10 |
paternally expressed 10 |
2240 |
0.29 |
| chr4_2418580_2419575 | 0.17 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
1259 |
0.44 |
| chr7_151106792_151107040 | 0.17 |
WDR86 |
WD repeat domain 86 |
190 |
0.8 |
| chr2_149633771_149633995 | 0.17 |
KIF5C |
kinesin family member 5C |
1064 |
0.51 |
| chr17_60215726_60216261 | 0.17 |
ENSG00000207123 |
. |
16312 |
0.22 |
| chr19_3062101_3062252 | 0.17 |
AES |
amino-terminal enhancer of split |
379 |
0.78 |
| chr8_23583978_23584470 | 0.16 |
RP11-175E9.1 |
|
19570 |
0.15 |
| chr4_80603993_80604144 | 0.16 |
OR7E94P |
olfactory receptor, family 7, subfamily E, member 94 pseudogene |
94784 |
0.09 |
| chr1_244893304_244893776 | 0.16 |
DESI2 |
desumoylating isopeptidase 2 |
76487 |
0.09 |
| chr3_44100057_44100240 | 0.16 |
ENSG00000252980 |
. |
12431 |
0.26 |
| chr7_29846053_29846716 | 0.16 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
282 |
0.94 |
| chr4_111532746_111532897 | 0.16 |
PITX2 |
paired-like homeodomain 2 |
11386 |
0.28 |
| chr6_88875424_88876640 | 0.16 |
CNR1 |
cannabinoid receptor 1 (brain) |
46 |
0.99 |
| chr7_106685641_106686008 | 0.16 |
PRKAR2B |
protein kinase, cAMP-dependent, regulatory, type II, beta |
730 |
0.74 |
| chrX_153686737_153687403 | 0.16 |
PLXNA3 |
plexin A3 |
449 |
0.6 |
| chr2_208635896_208636047 | 0.16 |
FZD5 |
frizzled family receptor 5 |
1684 |
0.3 |
| chr2_131792690_131793026 | 0.16 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
5992 |
0.24 |
| chr4_150999769_151000790 | 0.16 |
DCLK2 |
doublecortin-like kinase 2 |
99 |
0.98 |
| chr3_126077020_126077171 | 0.16 |
KLF15 |
Kruppel-like factor 15 |
810 |
0.63 |
| chr10_74513820_74513971 | 0.16 |
RP11-354E23.5 |
|
12423 |
0.18 |
| chr7_30721934_30722512 | 0.15 |
CRHR2 |
corticotropin releasing hormone receptor 2 |
91 |
0.98 |
| chr8_145489549_145489796 | 0.15 |
SCXA |
scleraxis homolog A (mouse) |
877 |
0.39 |
| chr8_145320438_145320697 | 0.15 |
SCXB |
scleraxis homolog B (mouse) |
950 |
0.47 |
| chr21_27011889_27012538 | 0.15 |
JAM2 |
junctional adhesion molecule 2 |
303 |
0.91 |
| chr2_27342148_27342407 | 0.15 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
282 |
0.79 |
| chr2_101435204_101435829 | 0.15 |
NPAS2 |
neuronal PAS domain protein 2 |
1098 |
0.54 |
| chr4_53727868_53728349 | 0.15 |
RASL11B |
RAS-like, family 11, member B |
349 |
0.9 |
| chr19_45675642_45675793 | 0.15 |
TRAPPC6A |
trafficking protein particle complex 6A |
5768 |
0.11 |
| chr22_29702254_29703430 | 0.15 |
GAS2L1 |
growth arrest-specific 2 like 1 |
171 |
0.91 |
| chr16_75284494_75285397 | 0.15 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
100 |
0.96 |
| chr9_137967884_137968035 | 0.15 |
OLFM1 |
olfactomedin 1 |
447 |
0.88 |
| chr4_52917683_52918097 | 0.15 |
SPATA18 |
spermatogenesis associated 18 |
308 |
0.91 |
| chr4_68904177_68904377 | 0.14 |
TMPRSS11GP |
transmembrane protease, serine 11G, pseudogene |
22791 |
0.16 |
| chr22_45054428_45054579 | 0.14 |
PRR5 |
proline rich 5 (renal) |
10090 |
0.23 |
| chr19_8372502_8373304 | 0.14 |
CD320 |
CD320 molecule |
331 |
0.8 |
| chr5_76372662_76373578 | 0.14 |
ENSG00000238961 |
. |
3276 |
0.17 |
| chr3_134093862_134094013 | 0.14 |
AMOTL2 |
angiomotin like 2 |
177 |
0.96 |
| chr9_131183467_131183728 | 0.14 |
CERCAM |
cerebral endothelial cell adhesion molecule |
837 |
0.44 |
| chr6_134210328_134210483 | 0.14 |
TCF21 |
transcription factor 21 |
68 |
0.76 |
| chr7_130578978_130579129 | 0.14 |
ENSG00000226380 |
. |
16755 |
0.25 |
| chr19_29590040_29590191 | 0.14 |
ENSG00000252272 |
. |
97538 |
0.08 |
| chr19_5294712_5294863 | 0.14 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
1524 |
0.49 |
| chr9_72286759_72287117 | 0.14 |
APBA1 |
amyloid beta (A4) precursor protein-binding, family A, member 1 |
284 |
0.94 |
| chr5_158518760_158518911 | 0.14 |
EBF1 |
early B-cell factor 1 |
7866 |
0.26 |
| chr14_96342973_96343124 | 0.14 |
ENSG00000263357 |
. |
25121 |
0.23 |
| chr19_46270942_46271877 | 0.14 |
AC074212.5 |
|
236 |
0.49 |
| chr17_6458496_6458984 | 0.14 |
PITPNM3 |
PITPNM family member 3 |
1040 |
0.49 |
| chrX_57618390_57619431 | 0.14 |
ZXDB |
zinc finger, X-linked, duplicated B |
641 |
0.84 |
| chr2_56150483_56150911 | 0.14 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
214 |
0.94 |
| chr3_8810545_8810722 | 0.14 |
OXTR |
oxytocin receptor |
270 |
0.88 |
| chr7_150758512_150758663 | 0.14 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
89 |
0.92 |
| chr1_1448548_1448833 | 0.14 |
ATAD3A |
ATPase family, AAA domain containing 3A |
780 |
0.47 |
| chr4_1075998_1076254 | 0.14 |
RNF212 |
ring finger protein 212 |
31188 |
0.1 |
| chr12_121648551_121648807 | 0.14 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
711 |
0.67 |
| chr7_55087572_55088165 | 0.14 |
EGFR |
epidermal growth factor receptor |
1057 |
0.68 |
| chr1_29448339_29449190 | 0.14 |
TMEM200B |
transmembrane protein 200B |
249 |
0.93 |
| chr18_46149088_46149239 | 0.14 |
ENSG00000266276 |
. |
47808 |
0.15 |
| chr7_102555257_102555409 | 0.13 |
LRRC17 |
leucine rich repeat containing 17 |
1881 |
0.34 |
| chr16_80972190_80972544 | 0.13 |
CMC2 |
C-x(9)-C motif containing 2 |
59897 |
0.09 |
| chr16_2198732_2199283 | 0.13 |
RAB26 |
RAB26, member RAS oncogene family |
382 |
0.61 |
| chr12_132313557_132314091 | 0.13 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
739 |
0.63 |
| chr17_48280882_48281058 | 0.13 |
COL1A1 |
collagen, type I, alpha 1 |
1977 |
0.21 |
| chr5_172140268_172140466 | 0.13 |
CTB-79E8.2 |
|
15644 |
0.15 |
| chr9_130739589_130739875 | 0.13 |
FAM102A |
family with sequence similarity 102, member A |
3060 |
0.15 |
| chr14_105877952_105878277 | 0.13 |
RP11-521B24.3 |
|
7962 |
0.12 |
| chr7_110732102_110732377 | 0.13 |
LRRN3 |
leucine rich repeat neuronal 3 |
1140 |
0.59 |
| chr16_31548902_31549248 | 0.13 |
AHSP |
alpha hemoglobin stabilizing protein |
9777 |
0.11 |
| chr19_30156409_30157288 | 0.13 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
440 |
0.86 |
| chr12_96183320_96183471 | 0.13 |
NTN4 |
netrin 4 |
331 |
0.86 |
| chr7_1578236_1578521 | 0.13 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
34 |
0.97 |
| chr1_165345218_165345646 | 0.13 |
LMX1A |
LIM homeobox transcription factor 1, alpha |
19480 |
0.23 |
| chr9_4490575_4491197 | 0.13 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
442 |
0.88 |
| chr10_85954746_85954956 | 0.13 |
CDHR1 |
cadherin-related family member 1 |
437 |
0.79 |
| chr19_51161641_51162497 | 0.13 |
C19orf81 |
chromosome 19 open reading frame 81 |
9024 |
0.12 |
| chr17_74025581_74025732 | 0.13 |
EVPL |
envoplakin |
2123 |
0.19 |
| chr1_56656926_56657077 | 0.13 |
ENSG00000223307 |
. |
186089 |
0.03 |
| chr1_85248095_85248299 | 0.13 |
ENSG00000251899 |
. |
10685 |
0.25 |
| chr13_40176285_40177298 | 0.13 |
LHFP |
lipoma HMGIC fusion partner |
517 |
0.82 |
| chr4_187025490_187026623 | 0.13 |
FAM149A |
family with sequence similarity 149, member A |
175 |
0.94 |
| chr6_148664293_148664728 | 0.13 |
SASH1 |
SAM and SH3 domain containing 1 |
781 |
0.74 |
| chr10_52751577_52751944 | 0.13 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
642 |
0.8 |
| chr10_17495370_17496061 | 0.13 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
614 |
0.77 |
| chr21_36042499_36043134 | 0.13 |
CLIC6 |
chloride intracellular channel 6 |
1128 |
0.59 |
| chr17_75428865_75429016 | 0.13 |
SEPT9 |
septin 9 |
5902 |
0.16 |
| chr12_123559968_123560341 | 0.13 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
488 |
0.8 |
| chr8_131455067_131455631 | 0.13 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
557 |
0.87 |
| chr5_41925427_41926188 | 0.13 |
FBXO4 |
F-box protein 4 |
404 |
0.85 |
| chr8_37555995_37556200 | 0.13 |
ZNF703 |
zinc finger protein 703 |
2828 |
0.18 |
| chr14_22955963_22956254 | 0.13 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
63 |
0.95 |
| chr12_8852377_8852528 | 0.13 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
10 |
0.98 |
| chrX_132092102_132092291 | 0.13 |
HS6ST2 |
heparan sulfate 6-O-sulfotransferase 2 |
852 |
0.73 |
| chr19_50861631_50862161 | 0.13 |
NAPSA |
napsin A aspartic peptidase |
6986 |
0.09 |
| chr10_22764645_22765757 | 0.13 |
RP11-301N24.3 |
|
115100 |
0.05 |
| chr7_5637806_5638004 | 0.12 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
3588 |
0.2 |
| chr17_38600498_38600710 | 0.12 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
891 |
0.5 |
| chr10_35926379_35926659 | 0.12 |
ENSG00000264780 |
. |
3661 |
0.21 |
| chr10_3109322_3109496 | 0.12 |
PFKP |
phosphofructokinase, platelet |
303 |
0.94 |
| chr6_43043530_43043730 | 0.12 |
PTK7 |
protein tyrosine kinase 7 |
376 |
0.67 |
| chr14_33407702_33407902 | 0.12 |
NPAS3 |
neuronal PAS domain protein 3 |
647 |
0.84 |
| chr9_133320745_133320993 | 0.12 |
ASS1 |
argininosuccinate synthase 1 |
490 |
0.76 |
| chr2_217500083_217500256 | 0.12 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
2051 |
0.31 |
| chrX_109675525_109675676 | 0.12 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
7861 |
0.23 |
| chr6_127836765_127837772 | 0.12 |
SOGA3 |
SOGA family member 3 |
491 |
0.85 |
| chr12_113670561_113670712 | 0.12 |
TPCN1 |
two pore segment channel 1 |
7587 |
0.13 |
| chr11_75943170_75943321 | 0.12 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
21465 |
0.18 |
| chr17_80054957_80055667 | 0.12 |
FASN |
fatty acid synthase |
896 |
0.37 |
| chr2_63817542_63817693 | 0.12 |
MDH1 |
malate dehydrogenase 1, NAD (soluble) |
1158 |
0.47 |
| chr10_98272514_98273588 | 0.12 |
TLL2 |
tolloid-like 2 |
617 |
0.79 |
| chr2_26785075_26785548 | 0.12 |
C2orf70 |
chromosome 2 open reading frame 70 |
139 |
0.96 |
| chr9_35649649_35650491 | 0.12 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
867 |
0.32 |
| chr1_32717802_32717953 | 0.12 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1002 |
0.32 |
| chr17_38614576_38614749 | 0.12 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
14949 |
0.13 |
| chr16_67197260_67198336 | 0.12 |
HSF4 |
heat shock transcription factor 4 |
510 |
0.52 |
| chr17_80355063_80355214 | 0.12 |
RP13-20L14.4 |
|
4120 |
0.11 |
| chr18_11571192_11571343 | 0.12 |
NPIPB1P |
nuclear pore complex interacting protein family, member B1, pseudogene |
68193 |
0.13 |
| chr2_74643272_74643423 | 0.12 |
C2orf81 |
chromosome 2 open reading frame 81 |
363 |
0.72 |
| chr17_71639222_71639967 | 0.12 |
SDK2 |
sidekick cell adhesion molecule 2 |
633 |
0.73 |
| chr1_85527614_85527975 | 0.12 |
WDR63 |
WD repeat domain 63 |
201 |
0.94 |
| chr10_79398606_79398757 | 0.12 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
328 |
0.92 |
| chr7_64029651_64030186 | 0.12 |
ZNF680 |
zinc finger protein 680 |
6434 |
0.3 |
| chr4_85419328_85419920 | 0.12 |
NKX6-1 |
NK6 homeobox 1 |
21 |
0.99 |
| chr6_72596608_72597181 | 0.12 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
167 |
0.98 |
| chr14_77227576_77228292 | 0.12 |
VASH1 |
vasohibin 1 |
598 |
0.75 |
| chr20_56754532_56754733 | 0.12 |
C20orf85 |
chromosome 20 open reading frame 85 |
28672 |
0.17 |
| chr7_128430672_128431008 | 0.12 |
CCDC136 |
coiled-coil domain containing 136 |
29 |
0.96 |
| chr21_47518056_47518267 | 0.12 |
COL6A2 |
collagen, type VI, alpha 2 |
128 |
0.89 |
| chr22_41957116_41957349 | 0.12 |
CSDC2 |
cold shock domain containing C2, RNA binding |
465 |
0.74 |
| chr22_20905991_20906349 | 0.12 |
MED15 |
mediator complex subunit 15 |
593 |
0.67 |
| chr22_28838455_28839140 | 0.12 |
ENSG00000199797 |
. |
139055 |
0.04 |
| chr19_13136060_13136211 | 0.12 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
324 |
0.8 |
| chr4_153168052_153168203 | 0.11 |
ENSG00000244544 |
. |
17470 |
0.2 |
| chr11_45201780_45202121 | 0.11 |
PRDM11 |
PR domain containing 11 |
1478 |
0.49 |
| chr4_184021252_184021496 | 0.11 |
WWC2 |
WW and C2 domain containing 2 |
729 |
0.73 |
| chr2_132218578_132219165 | 0.11 |
TUBA3D |
tubulin, alpha 3d |
14795 |
0.18 |
| chr1_147717990_147718807 | 0.11 |
ENSG00000199879 |
. |
17353 |
0.18 |
| chr4_113739356_113739869 | 0.11 |
ANK2 |
ankyrin 2, neuronal |
347 |
0.9 |
| chr14_69727713_69728275 | 0.11 |
GALNT16 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
986 |
0.63 |
| chr11_1892431_1892936 | 0.11 |
LSP1 |
lymphocyte-specific protein 1 |
567 |
0.59 |
| chr17_34121901_34122637 | 0.11 |
MMP28 |
matrix metallopeptidase 28 |
442 |
0.69 |
| chr17_71641526_71641788 | 0.11 |
SDK2 |
sidekick cell adhesion molecule 2 |
1429 |
0.46 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.2 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.1 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0031620 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 0.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.0 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0010665 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.0 | 0.0 | GO:0051580 | positive regulation of dopamine secretion(GO:0033603) regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0060761 | negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |