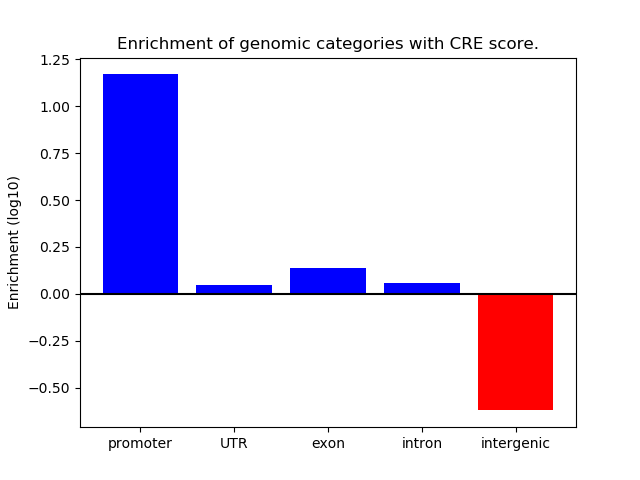
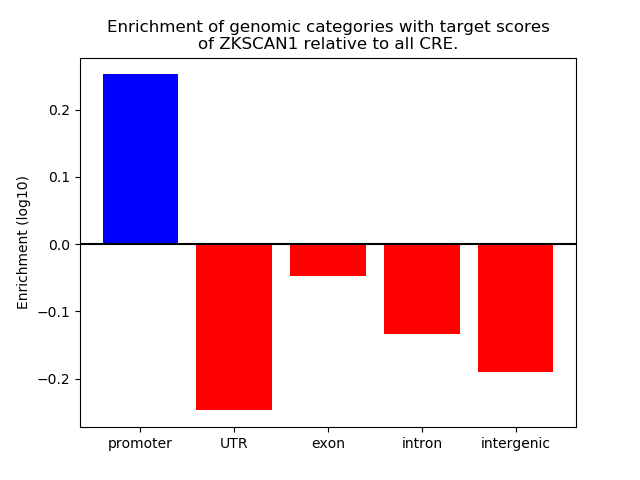
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
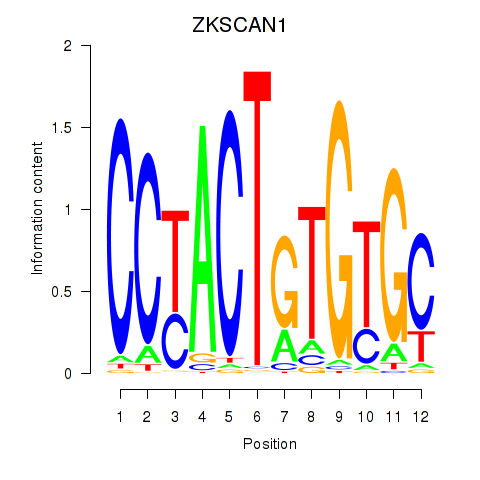
Results for ZKSCAN1
Z-value: 1.36
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | zinc finger with KRAB and SCAN domains 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
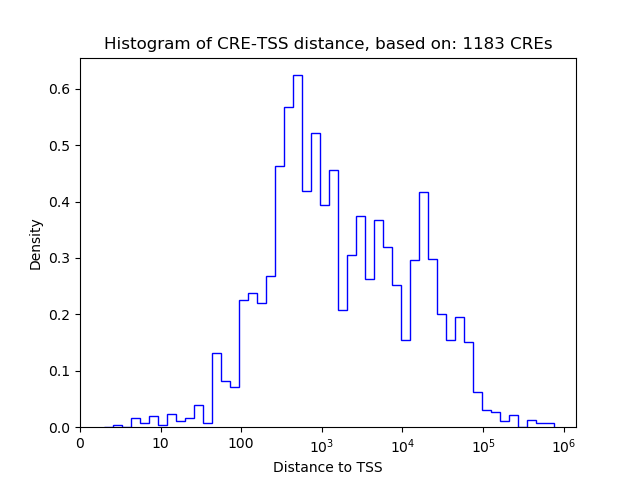
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_99595694_99595948 | ZKSCAN1 | 17383 | 0.083185 | 0.69 | 3.8e-02 | Click! |
| chr7_99604623_99604774 | ZKSCAN1 | 8506 | 0.094208 | 0.58 | 9.9e-02 | Click! |
| chr7_99613078_99613341 | ZKSCAN1 | 5 | 0.952124 | 0.50 | 1.7e-01 | Click! |
| chr7_99615127_99615278 | ZKSCAN1 | 757 | 0.457665 | -0.43 | 2.5e-01 | Click! |
| chr7_99620497_99620648 | ZKSCAN1 | 6127 | 0.097071 | -0.41 | 2.7e-01 | Click! |
Activity of the ZKSCAN1 motif across conditions
Conditions sorted by the z-value of the ZKSCAN1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
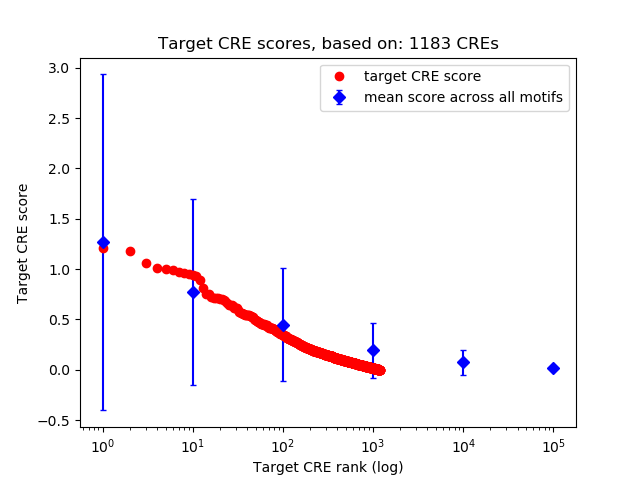
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_48770476_48770817 | 1.21 |
TMEM189 |
transmembrane protein 189 |
311 |
0.62 |
| chr9_123687593_123687825 | 1.18 |
TRAF1 |
TNF receptor-associated factor 1 |
3338 |
0.27 |
| chr22_44577668_44577918 | 1.06 |
PARVG |
parvin, gamma |
52 |
0.99 |
| chr19_10342329_10342480 | 1.01 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
442 |
0.39 |
| chr6_42015554_42016199 | 1.00 |
CCND3 |
cyclin D3 |
548 |
0.7 |
| chr19_16254302_16254453 | 0.99 |
HSH2D |
hematopoietic SH2 domain containing |
118 |
0.94 |
| chr6_159465090_159465296 | 0.98 |
TAGAP |
T-cell activation RhoGTPase activating protein |
857 |
0.62 |
| chr11_46367119_46367443 | 0.96 |
DGKZ |
diacylglycerol kinase, zeta |
294 |
0.88 |
| chr8_98806681_98806832 | 0.96 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
18679 |
0.16 |
| chr7_949432_949583 | 0.95 |
ADAP1 |
ArfGAP with dual PH domains 1 |
4793 |
0.16 |
| chr1_198611578_198611729 | 0.93 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
3361 |
0.31 |
| chr20_47436352_47436503 | 0.90 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
7993 |
0.28 |
| chr5_131762557_131762944 | 0.81 |
AC116366.5 |
|
344 |
0.83 |
| chr11_128343021_128343263 | 0.76 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
32147 |
0.21 |
| chr2_240219542_240220118 | 0.75 |
ENSG00000265215 |
. |
7327 |
0.19 |
| chr6_36922069_36922220 | 0.72 |
PI16 |
peptidase inhibitor 16 |
65 |
0.97 |
| chr20_31125041_31125259 | 0.72 |
C20orf112 |
chromosome 20 open reading frame 112 |
950 |
0.54 |
| chr19_13215215_13215439 | 0.71 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1352 |
0.27 |
| chr3_111851637_111852199 | 0.71 |
GCSAM |
germinal center-associated, signaling and motility |
154 |
0.73 |
| chr11_67184219_67184479 | 0.70 |
CARNS1 |
carnosine synthase 1 |
792 |
0.34 |
| chr15_75493595_75493850 | 0.70 |
C15orf39 |
chromosome 15 open reading frame 39 |
496 |
0.7 |
| chr1_208046696_208046851 | 0.70 |
CD34 |
CD34 molecule |
17064 |
0.25 |
| chr19_3178230_3178381 | 0.69 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
431 |
0.74 |
| chr19_10831379_10831550 | 0.66 |
DNM2 |
dynamin 2 |
2227 |
0.18 |
| chr1_17682926_17683077 | 0.64 |
AC004824.2 |
Uncharacterized protein |
6931 |
0.18 |
| chr13_99911056_99911338 | 0.64 |
GPR18 |
G protein-coupled receptor 18 |
515 |
0.8 |
| chr12_125424789_125424964 | 0.64 |
UBC |
ubiquitin C |
22962 |
0.14 |
| chr2_10444029_10444225 | 0.63 |
HPCAL1 |
hippocalcin-like 1 |
301 |
0.89 |
| chr5_71604472_71604655 | 0.61 |
ENSG00000244748 |
. |
5952 |
0.2 |
| chr6_165723730_165723881 | 0.61 |
C6orf118 |
chromosome 6 open reading frame 118 |
709 |
0.81 |
| chr19_18113119_18113338 | 0.61 |
ARRDC2 |
arrestin domain containing 2 |
1287 |
0.37 |
| chr17_36858727_36858878 | 0.60 |
ENSG00000265930 |
. |
218 |
0.82 |
| chr19_35632381_35632614 | 0.58 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
1488 |
0.2 |
| chr6_52929771_52930181 | 0.56 |
FBXO9 |
F-box protein 9 |
180 |
0.92 |
| chr12_50902852_50903181 | 0.56 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
4126 |
0.27 |
| chr15_41850352_41850568 | 0.55 |
TYRO3 |
TYRO3 protein tyrosine kinase |
587 |
0.7 |
| chr5_134240090_134240263 | 0.55 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
420 |
0.81 |
| chr7_30766706_30766866 | 0.55 |
INMT |
indolethylamine N-methyltransferase |
24965 |
0.16 |
| chr19_2489456_2489664 | 0.54 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
13206 |
0.14 |
| chr2_144005287_144005438 | 0.54 |
RP11-190J23.1 |
|
75621 |
0.11 |
| chr15_45002906_45003073 | 0.54 |
B2M |
beta-2-microglobulin |
686 |
0.62 |
| chr11_79212076_79212227 | 0.54 |
TENM4 |
teneurin transmembrane protein 4 |
60456 |
0.14 |
| chrX_23970840_23971181 | 0.53 |
CXorf58 |
chromosome X open reading frame 58 |
42510 |
0.13 |
| chr9_126101480_126101665 | 0.53 |
CRB2 |
crumbs homolog 2 (Drosophila) |
16877 |
0.22 |
| chr1_173445561_173445725 | 0.53 |
PRDX6 |
peroxiredoxin 6 |
762 |
0.68 |
| chr15_40731920_40732071 | 0.52 |
BAHD1 |
bromo adjacent homology domain containing 1 |
75 |
0.94 |
| chr1_28199781_28200360 | 0.52 |
THEMIS2 |
thymocyte selection associated family member 2 |
991 |
0.4 |
| chr13_52411655_52411806 | 0.50 |
CCDC70 |
coiled-coil domain containing 70 |
24387 |
0.17 |
| chr19_15574852_15575181 | 0.50 |
RASAL3 |
RAS protein activator like 3 |
366 |
0.8 |
| chr9_132647660_132648047 | 0.49 |
FNBP1 |
formin binding protein 1 |
33736 |
0.12 |
| chr17_40439619_40439799 | 0.49 |
STAT5A |
signal transducer and activator of transcription 5A |
144 |
0.93 |
| chr8_37755503_37755707 | 0.49 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
1367 |
0.34 |
| chr4_1210592_1210772 | 0.48 |
CTBP1 |
C-terminal binding protein 1 |
2432 |
0.21 |
| chr12_31811913_31812109 | 0.48 |
METTL20 |
methyltransferase like 20 |
110 |
0.96 |
| chr3_161261196_161261347 | 0.47 |
OTOL1 |
otolin 1 |
46675 |
0.2 |
| chr16_67062127_67062421 | 0.46 |
CBFB |
core-binding factor, beta subunit |
745 |
0.53 |
| chr16_27326766_27327107 | 0.46 |
IL4R |
interleukin 4 receptor |
1623 |
0.4 |
| chr19_6801178_6801329 | 0.46 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
17124 |
0.13 |
| chr14_95963717_95963868 | 0.45 |
RP11-1070N10.3 |
Uncharacterized protein |
18681 |
0.14 |
| chr9_115096689_115097200 | 0.45 |
PTBP3 |
polypyrimidine tract binding protein 3 |
997 |
0.57 |
| chr1_6659044_6659355 | 0.45 |
KLHL21 |
kelch-like family member 21 |
677 |
0.56 |
| chr15_60771714_60772006 | 0.45 |
RP11-219B17.1 |
|
441 |
0.52 |
| chr4_109087263_109087414 | 0.45 |
LEF1 |
lymphoid enhancer-binding factor 1 |
119 |
0.95 |
| chr4_2814595_2815029 | 0.44 |
SH3BP2 |
SH3-domain binding protein 2 |
781 |
0.66 |
| chr10_23384240_23384391 | 0.44 |
MSRB2 |
methionine sulfoxide reductase B2 |
120 |
0.97 |
| chr10_103539043_103539292 | 0.44 |
FGF8 |
fibroblast growth factor 8 (androgen-induced) |
3340 |
0.19 |
| chr7_86689559_86689710 | 0.44 |
KIAA1324L |
KIAA1324-like |
619 |
0.81 |
| chr19_2489761_2489912 | 0.43 |
ENSG00000252962 |
. |
13303 |
0.14 |
| chr17_73151022_73151194 | 0.43 |
HN1 |
hematological and neurological expressed 1 |
330 |
0.75 |
| chr15_77286829_77286980 | 0.42 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
522 |
0.79 |
| chr20_62198919_62199070 | 0.42 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
433 |
0.7 |
| chr6_79902044_79902327 | 0.41 |
HMGN3-AS1 |
HMGN3 antisense RNA 1 |
41250 |
0.16 |
| chr19_45258275_45258481 | 0.41 |
BCL3 |
B-cell CLL/lymphoma 3 |
3800 |
0.13 |
| chr22_24180306_24180495 | 0.41 |
DERL3 |
derlin 3 |
791 |
0.44 |
| chr19_8070956_8071107 | 0.41 |
ELAVL1 |
ELAV like RNA binding protein 1 |
488 |
0.72 |
| chr15_81593596_81593907 | 0.41 |
IL16 |
interleukin 16 |
1994 |
0.33 |
| chr19_42388528_42389094 | 0.41 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
296 |
0.84 |
| chr2_74611000_74611151 | 0.41 |
DCTN1 |
dynactin 1 |
1493 |
0.21 |
| chr9_131964929_131965166 | 0.41 |
RP11-247A12.2 |
|
1453 |
0.33 |
| chr1_55678186_55678720 | 0.41 |
USP24 |
ubiquitin specific peptidase 24 |
2309 |
0.34 |
| chr1_85871559_85871710 | 0.40 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
1454 |
0.45 |
| chr12_71167942_71168093 | 0.39 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
14745 |
0.28 |
| chr1_27849169_27849399 | 0.39 |
RP1-159A19.4 |
|
3032 |
0.23 |
| chr1_234613311_234613591 | 0.39 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
1398 |
0.49 |
| chr20_43279687_43280217 | 0.38 |
ADA |
adenosine deaminase |
380 |
0.81 |
| chr20_3998751_3998989 | 0.38 |
RNF24 |
ring finger protein 24 |
2641 |
0.31 |
| chr8_134511768_134511919 | 0.38 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
217 |
0.97 |
| chr13_36410557_36410708 | 0.37 |
DCLK1 |
doublecortin-like kinase 1 |
19147 |
0.29 |
| chrY_15590512_15590663 | 0.37 |
UTY |
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
958 |
0.72 |
| chr11_75063727_75063930 | 0.37 |
ARRB1 |
arrestin, beta 1 |
955 |
0.48 |
| chr6_157098499_157098696 | 0.37 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
466 |
0.78 |
| chr9_137856018_137856169 | 0.37 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
46284 |
0.11 |
| chr6_10529373_10529624 | 0.36 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
909 |
0.59 |
| chr7_155556612_155556872 | 0.36 |
RBM33 |
RNA binding motif protein 33 |
2508 |
0.35 |
| chr7_124289801_124290098 | 0.36 |
ENSG00000207214 |
. |
2176 |
0.46 |
| chr22_45680566_45680776 | 0.35 |
UPK3A |
uroplakin 3A |
192 |
0.94 |
| chrX_131622234_131622385 | 0.35 |
MBNL3 |
muscleblind-like splicing regulator 3 |
734 |
0.78 |
| chr7_100202948_100203247 | 0.35 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
1268 |
0.23 |
| chr9_139444261_139444412 | 0.35 |
RP11-611D20.2 |
|
754 |
0.45 |
| chr16_19729638_19729939 | 0.35 |
IQCK |
IQ motif containing K |
151 |
0.61 |
| chr2_75065127_75065278 | 0.35 |
HK2 |
hexokinase 2 |
2905 |
0.34 |
| chr13_41372571_41372761 | 0.34 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
5372 |
0.18 |
| chr19_35820197_35820463 | 0.34 |
CD22 |
CD22 molecule |
215 |
0.83 |
| chr19_45606414_45606793 | 0.33 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
10171 |
0.09 |
| chr5_118609514_118610204 | 0.33 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
5410 |
0.21 |
| chr1_42206292_42206443 | 0.33 |
ENSG00000264896 |
. |
18445 |
0.24 |
| chr1_87169604_87170085 | 0.33 |
RP4-612B15.3 |
|
301 |
0.59 |
| chr17_76772724_76772875 | 0.33 |
CYTH1 |
cytohesin 1 |
5555 |
0.2 |
| chr17_36858943_36859359 | 0.33 |
ENSG00000265930 |
. |
567 |
0.49 |
| chr2_71751488_71751639 | 0.32 |
DYSF |
dysferlin |
57731 |
0.15 |
| chr5_134735051_134735464 | 0.32 |
H2AFY |
H2A histone family, member Y |
55 |
0.79 |
| chr11_76091032_76091428 | 0.31 |
PRKRIR |
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
756 |
0.46 |
| chr8_66751055_66751459 | 0.31 |
PDE7A |
phosphodiesterase 7A |
274 |
0.95 |
| chr19_2092800_2093080 | 0.31 |
MOB3A |
MOB kinase activator 3A |
2765 |
0.15 |
| chr10_106089117_106089268 | 0.31 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
510 |
0.71 |
| chr11_75943170_75943321 | 0.31 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
21465 |
0.18 |
| chrX_78402769_78403059 | 0.31 |
GPR174 |
G protein-coupled receptor 174 |
23555 |
0.28 |
| chr5_169724277_169724686 | 0.31 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
750 |
0.71 |
| chr12_46063121_46063382 | 0.31 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
60197 |
0.15 |
| chr3_69590054_69590354 | 0.30 |
FRMD4B |
FERM domain containing 4B |
287 |
0.95 |
| chr16_79123602_79124194 | 0.30 |
RP11-556H2.3 |
|
49 |
0.98 |
| chr15_78327297_78327492 | 0.30 |
ENSG00000221476 |
. |
3479 |
0.18 |
| chr16_69346193_69346377 | 0.30 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
1026 |
0.35 |
| chr7_142498684_142498938 | 0.30 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
17680 |
0.17 |
| chr7_149411220_149411666 | 0.30 |
KRBA1 |
KRAB-A domain containing 1 |
429 |
0.87 |
| chr1_11332767_11332951 | 0.29 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
404 |
0.83 |
| chr12_120548897_120549070 | 0.29 |
RAB35 |
RAB35, member RAS oncogene family |
5557 |
0.17 |
| chr3_148803795_148804035 | 0.29 |
HLTF-AS1 |
HLTF antisense RNA 1 |
204 |
0.68 |
| chr22_33040047_33040198 | 0.29 |
ENSG00000251890 |
. |
8938 |
0.25 |
| chrX_109245290_109245568 | 0.29 |
TMEM164 |
transmembrane protein 164 |
434 |
0.88 |
| chr19_35494097_35494396 | 0.29 |
GRAMD1A |
GRAM domain containing 1A |
2886 |
0.14 |
| chr5_72731379_72731530 | 0.28 |
FOXD1 |
forkhead box D1 |
12898 |
0.21 |
| chrX_135578358_135578906 | 0.28 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
606 |
0.66 |
| chr19_39420489_39421004 | 0.28 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
27 |
0.86 |
| chr6_31240086_31240407 | 0.28 |
HLA-C |
major histocompatibility complex, class I, C |
383 |
0.81 |
| chr2_208007462_208007613 | 0.28 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
8676 |
0.2 |
| chr7_5571391_5571891 | 0.28 |
ACTB |
actin, beta |
1301 |
0.32 |
| chr8_142413039_142413204 | 0.28 |
CTD-3064M3.4 |
|
10447 |
0.12 |
| chr11_65379543_65379824 | 0.28 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
911 |
0.31 |
| chr1_230406303_230406454 | 0.28 |
RP5-956O18.2 |
|
2149 |
0.36 |
| chr17_43505837_43505988 | 0.28 |
ARHGAP27 |
Rho GTPase activating protein 27 |
1733 |
0.23 |
| chr17_33446349_33446789 | 0.28 |
RAD51D |
RAD51 paralog D |
4 |
0.88 |
| chr14_99727366_99727576 | 0.27 |
AL109767.1 |
|
1814 |
0.39 |
| chrX_47441424_47441613 | 0.27 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
194 |
0.91 |
| chr11_67981482_67982050 | 0.26 |
SUV420H1 |
suppressor of variegation 4-20 homolog 1 (Drosophila) |
471 |
0.83 |
| chr13_53484556_53484707 | 0.26 |
PCDH8 |
protocadherin 8 |
61856 |
0.13 |
| chr1_19812521_19812692 | 0.26 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
540 |
0.74 |
| chr5_170818755_170818906 | 0.26 |
NPM1 |
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
3945 |
0.18 |
| chr6_24925347_24925503 | 0.26 |
FAM65B |
family with sequence similarity 65, member B |
10763 |
0.23 |
| chrX_139006308_139006459 | 0.25 |
ENSG00000207633 |
. |
7 |
0.97 |
| chr19_42376193_42376392 | 0.25 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
4898 |
0.12 |
| chr15_65822500_65822707 | 0.25 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
153 |
0.94 |
| chr16_791373_791524 | 0.25 |
NARFL |
nuclear prelamin A recognition factor-like |
119 |
0.88 |
| chr19_4060134_4060285 | 0.25 |
CTD-2622I13.3 |
|
2538 |
0.17 |
| chr12_75875877_75876340 | 0.25 |
GLIPR1 |
GLI pathogenesis-related 1 |
1124 |
0.49 |
| chr1_111764570_111764791 | 0.25 |
CHI3L2 |
chitinase 3-like 2 |
4958 |
0.15 |
| chr15_75417454_75417605 | 0.24 |
ENSG00000252722 |
. |
3522 |
0.21 |
| chr9_131842660_131843064 | 0.24 |
DOLPP1 |
dolichyldiphosphatase 1 |
517 |
0.67 |
| chr20_43513806_43514102 | 0.24 |
YWHAB |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
363 |
0.83 |
| chr4_6942044_6942219 | 0.24 |
ENSG00000265953 |
. |
12885 |
0.15 |
| chr5_173202456_173202607 | 0.24 |
ENSG00000263401 |
. |
45450 |
0.18 |
| chr4_56814750_56814930 | 0.24 |
CEP135 |
centrosomal protein 135kDa |
197 |
0.95 |
| chrX_152864941_152865619 | 0.24 |
FAM58A |
family with sequence similarity 58, member A |
220 |
0.71 |
| chrX_39796396_39796688 | 0.24 |
ENSG00000263972 |
. |
99727 |
0.08 |
| chr20_826611_826818 | 0.23 |
FAM110A |
family with sequence similarity 110, member A |
947 |
0.64 |
| chr6_109560429_109560580 | 0.23 |
ENSG00000238474 |
. |
52057 |
0.12 |
| chr14_105635560_105635849 | 0.23 |
JAG2 |
jagged 2 |
543 |
0.75 |
| chr11_48018118_48018269 | 0.23 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
15914 |
0.19 |
| chr7_143072173_143072546 | 0.23 |
AC093673.5 |
|
5229 |
0.12 |
| chr8_1430917_1431068 | 0.23 |
DLGAP2 |
discs, large (Drosophila) homolog-associated protein 2 |
18540 |
0.28 |
| chr15_55569874_55570045 | 0.23 |
RAB27A |
RAB27A, member RAS oncogene family |
6852 |
0.2 |
| chr17_55332585_55332736 | 0.23 |
MSI2 |
musashi RNA-binding protein 2 |
552 |
0.84 |
| chr6_154570744_154570920 | 0.23 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
2016 |
0.49 |
| chr1_205409872_205410317 | 0.23 |
ENSG00000199059 |
. |
7432 |
0.17 |
| chr19_5294712_5294863 | 0.23 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
1524 |
0.49 |
| chr3_97438061_97438212 | 0.23 |
RP11-389G6.3 |
|
40231 |
0.17 |
| chr10_124713000_124713296 | 0.22 |
C10orf88 |
chromosome 10 open reading frame 88 |
771 |
0.61 |
| chr1_26561482_26561883 | 0.22 |
CEP85 |
centrosomal protein 85kDa |
989 |
0.41 |
| chr16_25269686_25269905 | 0.22 |
ZKSCAN2 |
zinc finger with KRAB and SCAN domains 2 |
543 |
0.78 |
| chr9_82317273_82317424 | 0.22 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
2350 |
0.47 |
| chr1_205256784_205256935 | 0.22 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
31530 |
0.13 |
| chr16_31520088_31520277 | 0.22 |
RP11-452L6.7 |
|
390 |
0.47 |
| chr15_70773947_70774098 | 0.22 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
220598 |
0.02 |
| chr12_4380598_4381153 | 0.22 |
CCND2 |
cyclin D2 |
2063 |
0.27 |
| chr22_38055772_38055923 | 0.22 |
PDXP |
pyridoxal (pyridoxine, vitamin B6) phosphatase |
1113 |
0.27 |
| chr7_157361164_157361701 | 0.22 |
ENSG00000207960 |
. |
5682 |
0.22 |
| chr14_32671845_32671996 | 0.22 |
ENSG00000202337 |
. |
555 |
0.52 |
| chr22_25003998_25004507 | 0.22 |
GGT1 |
gamma-glutamyltransferase 1 |
621 |
0.67 |
| chr8_131029198_131029390 | 0.22 |
FAM49B |
family with sequence similarity 49, member B |
81 |
0.97 |
| chr12_120966627_120966937 | 0.22 |
COQ5 |
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
161 |
0.91 |
| chr1_201476350_201476659 | 0.21 |
CSRP1 |
cysteine and glycine-rich protein 1 |
117 |
0.52 |
| chr12_125034711_125034862 | 0.21 |
NCOR2 |
nuclear receptor corepressor 2 |
14637 |
0.29 |
| chr1_16302650_16302808 | 0.21 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
102 |
0.95 |
| chr22_38082633_38082863 | 0.21 |
NOL12 |
nucleolar protein 12 |
418 |
0.71 |
| chr11_17497808_17497959 | 0.21 |
ABCC8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
509 |
0.81 |
| chr4_40241072_40241223 | 0.21 |
RHOH |
ras homolog family member H |
39183 |
0.16 |
| chr1_38312116_38312382 | 0.21 |
MTF1 |
metal-regulatory transcription factor 1 |
13043 |
0.09 |
| chr1_117079785_117079936 | 0.21 |
CD58 |
CD58 molecule |
7352 |
0.18 |
| chr2_70775363_70775514 | 0.21 |
TGFA |
transforming growth factor, alpha |
5184 |
0.25 |
| chr8_142427041_142427192 | 0.21 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
4891 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 0.8 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.2 | 0.5 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.1 | 0.4 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 0.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 0.3 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.4 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.1 | 0.3 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.1 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.3 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.1 | 0.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.5 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.3 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0071028 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0021955 | central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0090196 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0039694 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.0 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.9 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.9 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |