Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
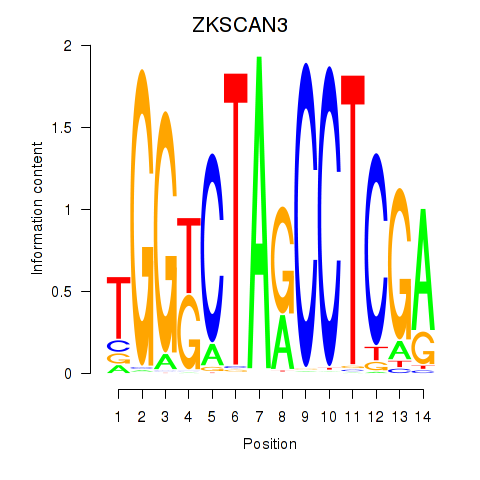
Results for ZKSCAN3
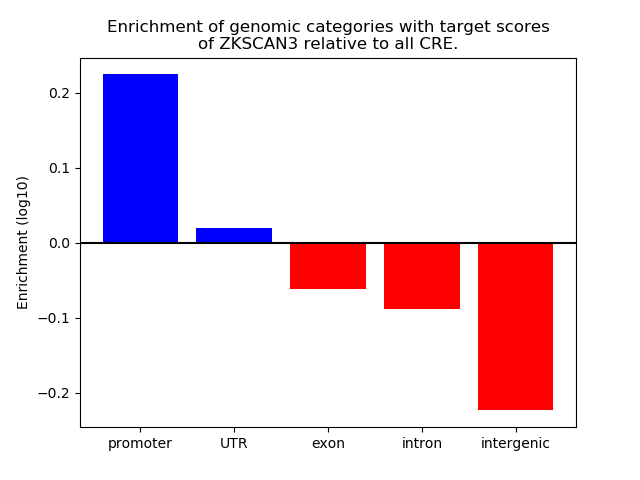
Z-value: 1.48
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.9 | zinc finger with KRAB and SCAN domains 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_28317524_28317690 | ZKSCAN3 | 84 | 0.960724 | 0.08 | 8.3e-01 | Click! |
| chr6_28317691_28318186 | ZKSCAN3 | 211 | 0.915228 | -0.04 | 9.1e-01 | Click! |
Activity of the ZKSCAN3 motif across conditions
Conditions sorted by the z-value of the ZKSCAN3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
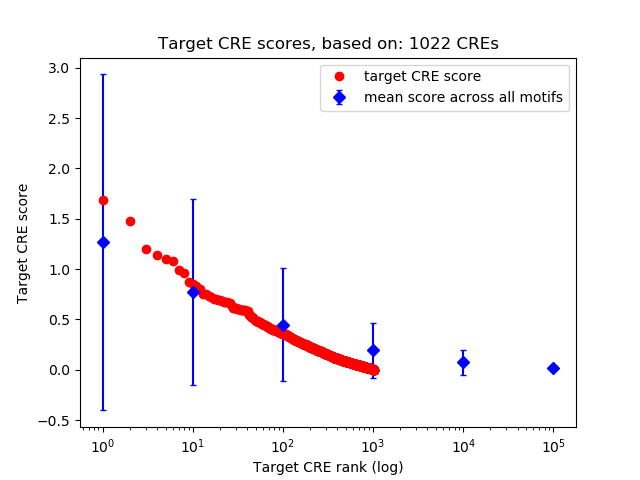
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_56408975_56409725 | 1.69 |
MIR142 |
microRNA 142 |
519 |
0.63 |
| chr17_76126911_76127107 | 1.48 |
TMC8 |
transmembrane channel-like 8 |
140 |
0.63 |
| chrX_128914916_128915218 | 1.20 |
SASH3 |
SAM and SH3 domain containing 3 |
1107 |
0.52 |
| chr5_118607055_118607314 | 1.15 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2735 |
0.27 |
| chr20_56193926_56194226 | 1.10 |
ZBP1 |
Z-DNA binding protein 1 |
1374 |
0.49 |
| chr14_76009252_76009829 | 1.08 |
BATF |
basic leucine zipper transcription factor, ATF-like |
20637 |
0.15 |
| chr1_1148809_1149188 | 1.00 |
TNFRSF4 |
tumor necrosis factor receptor superfamily, member 4 |
514 |
0.56 |
| chr11_3860110_3860464 | 0.96 |
RHOG |
ras homolog family member G |
1190 |
0.3 |
| chr4_6918537_6918793 | 0.88 |
TBC1D14 |
TBC1 domain family, member 14 |
6690 |
0.19 |
| chr2_136872491_136872642 | 0.85 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
1247 |
0.6 |
| chr16_70452884_70453127 | 0.84 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
18464 |
0.11 |
| chr20_826059_826564 | 0.80 |
FAM110A |
family with sequence similarity 110, member A |
544 |
0.82 |
| chr3_9821738_9821889 | 0.75 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
10137 |
0.09 |
| chr20_50158730_50159057 | 0.75 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
365 |
0.92 |
| chr2_169103559_169103719 | 0.73 |
STK39 |
serine threonine kinase 39 |
1012 |
0.7 |
| chr1_7763805_7764045 | 0.72 |
CAMTA1 |
calmodulin binding transcription activator 1 |
32543 |
0.17 |
| chr14_24640773_24641022 | 0.70 |
REC8 |
REC8 meiotic recombination protein |
165 |
0.84 |
| chr3_43332584_43332867 | 0.70 |
SNRK |
SNF related kinase |
4647 |
0.21 |
| chr1_211509571_211509738 | 0.70 |
TRAF5 |
TNF receptor-associated factor 5 |
9475 |
0.25 |
| chr3_127392819_127392970 | 0.69 |
ABTB1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
59 |
0.97 |
| chr13_30950896_30951047 | 0.68 |
KATNAL1 |
katanin p60 subunit A-like 1 |
69350 |
0.11 |
| chr18_21601315_21601466 | 0.68 |
TTC39C |
tetratricopeptide repeat domain 39C |
4792 |
0.19 |
| chr4_154448881_154449032 | 0.67 |
KIAA0922 |
KIAA0922 |
28092 |
0.22 |
| chr17_45812399_45812645 | 0.67 |
TBX21 |
T-box 21 |
1912 |
0.27 |
| chrY_22738367_22738527 | 0.66 |
EIF1AY |
eukaryotic translation initiation factor 1A, Y-linked |
767 |
0.8 |
| chr4_42657844_42658977 | 0.66 |
ATP8A1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
700 |
0.59 |
| chr9_20379288_20379439 | 0.64 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
3104 |
0.27 |
| chr17_33640471_33640814 | 0.62 |
SLFN11 |
schlafen family member 11 |
39508 |
0.12 |
| chr20_31069881_31070147 | 0.62 |
C20orf112 |
chromosome 20 open reading frame 112 |
1260 |
0.46 |
| chr1_62862864_62863015 | 0.61 |
USP1 |
ubiquitin specific peptidase 1 |
39029 |
0.17 |
| chr2_71292065_71292344 | 0.61 |
AC007040.8 |
|
331 |
0.79 |
| chr11_46140422_46140573 | 0.60 |
PHF21A |
PHD finger protein 21A |
841 |
0.64 |
| chr1_9690042_9690512 | 0.60 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
21513 |
0.15 |
| chr19_42703347_42703984 | 0.59 |
ENSG00000265122 |
. |
6435 |
0.1 |
| chr3_81692511_81692662 | 0.59 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
100194 |
0.09 |
| chr2_152326029_152326180 | 0.59 |
NEB |
nebulin |
33839 |
0.18 |
| chr14_94429340_94429491 | 0.59 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
5648 |
0.17 |
| chr14_78108377_78108627 | 0.59 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
25386 |
0.16 |
| chr8_22876543_22876783 | 0.59 |
RP11-875O11.1 |
|
185 |
0.92 |
| chr6_30458735_30458886 | 0.59 |
HLA-E |
major histocompatibility complex, class I, E |
1566 |
0.31 |
| chr9_71650379_71650667 | 0.58 |
FXN |
frataxin |
44 |
0.98 |
| chr9_129485287_129485658 | 0.55 |
ENSG00000266403 |
. |
4860 |
0.26 |
| chr7_75267461_75267701 | 0.54 |
HIP1 |
huntingtin interacting protein 1 |
26420 |
0.17 |
| chr11_58291916_58292352 | 0.53 |
OR5B21 |
olfactory receptor, family 5, subfamily B, member 21 |
16556 |
0.16 |
| chr2_103035996_103036147 | 0.52 |
IL18RAP |
interleukin 18 receptor accessory protein |
75 |
0.96 |
| chr16_14379797_14380077 | 0.52 |
ENSG00000201075 |
. |
10575 |
0.18 |
| chr2_198063557_198063736 | 0.51 |
ANKRD44 |
ankyrin repeat domain 44 |
884 |
0.43 |
| chr10_97516050_97516539 | 0.51 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
580 |
0.8 |
| chr10_121276029_121276279 | 0.49 |
RGS10 |
regulator of G-protein signaling 10 |
10835 |
0.24 |
| chr12_66585349_66585521 | 0.49 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
2398 |
0.25 |
| chr9_34122956_34123107 | 0.48 |
DCAF12 |
DDB1 and CUL4 associated factor 12 |
2694 |
0.22 |
| chr10_115613974_115614183 | 0.48 |
DCLRE1A |
DNA cross-link repair 1A |
64 |
0.82 |
| chr1_226926024_226926994 | 0.48 |
ITPKB |
inositol-trisphosphate 3-kinase B |
355 |
0.91 |
| chr12_49374311_49374525 | 0.48 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
2020 |
0.12 |
| chrX_153672104_153672338 | 0.48 |
FAM50A |
family with sequence similarity 50, member A |
252 |
0.79 |
| chr8_66752287_66752671 | 0.47 |
PDE7A |
phosphodiesterase 7A |
1264 |
0.6 |
| chr20_58515606_58515958 | 0.46 |
FAM217B |
family with sequence similarity 217, member B |
288 |
0.62 |
| chr3_47028874_47029030 | 0.45 |
CCDC12 |
coiled-coil domain containing 12 |
5480 |
0.19 |
| chr13_49787907_49788058 | 0.45 |
MLNR |
motilin receptor |
6492 |
0.24 |
| chr21_47971505_47971656 | 0.45 |
ENSG00000272283 |
. |
15871 |
0.17 |
| chr2_191744870_191745300 | 0.45 |
GLS |
glutaminase |
468 |
0.79 |
| chr21_34395985_34396161 | 0.45 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
2080 |
0.33 |
| chr3_122512790_122513032 | 0.44 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
240 |
0.82 |
| chr22_50311854_50312054 | 0.44 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
139 |
0.51 |
| chr19_45595083_45595471 | 0.44 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
941 |
0.32 |
| chr2_231533080_231533231 | 0.44 |
CAB39 |
calcium binding protein 39 |
44405 |
0.14 |
| chr12_123372500_123372784 | 0.43 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
2064 |
0.29 |
| chr6_157970217_157970368 | 0.42 |
ENSG00000266617 |
. |
20128 |
0.23 |
| chr1_149294050_149294201 | 0.42 |
ENSG00000252105 |
. |
7803 |
0.17 |
| chr4_185741986_185742137 | 0.42 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
5127 |
0.2 |
| chr1_185083987_185084138 | 0.41 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
24630 |
0.18 |
| chr11_117198864_117199680 | 0.41 |
CEP164 |
centrosomal protein 164kDa |
453 |
0.5 |
| chr9_139000986_139001191 | 0.41 |
C9orf69 |
chromosome 9 open reading frame 69 |
9032 |
0.19 |
| chr15_41624315_41624466 | 0.41 |
OIP5 |
Opa interacting protein 5 |
295 |
0.65 |
| chr2_85107193_85107344 | 0.40 |
TRABD2A |
TraB domain containing 2A |
938 |
0.57 |
| chr16_23689854_23690079 | 0.40 |
PLK1 |
polo-like kinase 1 |
177 |
0.92 |
| chr14_50366151_50366383 | 0.40 |
ENSG00000251929 |
. |
2401 |
0.2 |
| chr6_158980808_158981008 | 0.40 |
TMEM181 |
transmembrane protein 181 |
23440 |
0.19 |
| chr2_10548605_10548929 | 0.40 |
HPCAL1 |
hippocalcin-like 1 |
11380 |
0.19 |
| chr17_29158612_29158825 | 0.39 |
ATAD5 |
ATPase family, AAA domain containing 5 |
270 |
0.86 |
| chr1_180982153_180982304 | 0.39 |
STX6 |
syntaxin 6 |
9777 |
0.16 |
| chr16_84801882_84802033 | 0.39 |
USP10 |
ubiquitin specific peptidase 10 |
84 |
0.98 |
| chr12_8125010_8125161 | 0.39 |
ENSG00000206636 |
. |
3770 |
0.2 |
| chr7_35731550_35731936 | 0.39 |
HERPUD2 |
HERPUD family member 2 |
2433 |
0.31 |
| chr8_21776103_21776686 | 0.39 |
XPO7 |
exportin 7 |
786 |
0.64 |
| chr7_150178540_150178691 | 0.38 |
GIMAP8 |
GTPase, IMAP family member 8 |
30897 |
0.13 |
| chr19_23427286_23427437 | 0.38 |
ZNF724P |
zinc finger protein 724, pseudogene |
5801 |
0.26 |
| chr8_37754904_37755445 | 0.38 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
1798 |
0.27 |
| chrX_144516660_144516811 | 0.38 |
SPANXN1 |
SPANX family, member N1 |
188387 |
0.03 |
| chr16_27326766_27327107 | 0.38 |
IL4R |
interleukin 4 receptor |
1623 |
0.4 |
| chr12_77060372_77060523 | 0.37 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
96921 |
0.08 |
| chr15_91428804_91428955 | 0.37 |
FES |
feline sarcoma oncogene |
603 |
0.56 |
| chr2_42333200_42333351 | 0.37 |
PKDCC |
protein kinase domain containing, cytoplasmic |
58115 |
0.11 |
| chr19_7198675_7198826 | 0.37 |
INSR |
insulin receptor |
30761 |
0.15 |
| chr6_90538863_90539246 | 0.37 |
ENSG00000238747 |
. |
6040 |
0.17 |
| chr5_14581323_14581785 | 0.36 |
FAM105A |
family with sequence similarity 105, member A |
330 |
0.92 |
| chr11_125773182_125774068 | 0.36 |
PUS3 |
pseudouridylate synthase 3 |
509 |
0.49 |
| chr7_26564902_26565053 | 0.36 |
KIAA0087 |
KIAA0087 |
13430 |
0.27 |
| chr12_113573231_113573417 | 0.36 |
RASAL1 |
RAS protein activator like 1 (GAP1 like) |
15 |
0.96 |
| chr12_7064003_7064154 | 0.36 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
3540 |
0.07 |
| chr15_83777071_83777287 | 0.36 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
716 |
0.58 |
| chr7_92464516_92465198 | 0.35 |
AC002454.1 |
|
940 |
0.44 |
| chr17_75451832_75452118 | 0.35 |
SEPT9 |
septin 9 |
473 |
0.77 |
| chr1_153204493_153204644 | 0.35 |
PRR9 |
proline rich 9 |
14508 |
0.12 |
| chr12_104682714_104682980 | 0.35 |
TXNRD1 |
thioredoxin reductase 1 |
174 |
0.95 |
| chr8_53851957_53852159 | 0.35 |
NPBWR1 |
neuropeptides B/W receptor 1 |
1067 |
0.63 |
| chr12_105379814_105380296 | 0.35 |
C12orf45 |
chromosome 12 open reading frame 45 |
33 |
0.98 |
| chr11_73091522_73091673 | 0.35 |
RELT |
RELT tumor necrosis factor receptor |
3884 |
0.17 |
| chr19_2702225_2702378 | 0.34 |
GNG7 |
guanine nucleotide binding protein (G protein), gamma 7 |
406 |
0.76 |
| chr19_9649363_9649620 | 0.34 |
ZNF426 |
zinc finger protein 426 |
188 |
0.94 |
| chr1_149679650_149680105 | 0.34 |
FAM231D |
family with sequence similarity 231, member D |
3899 |
0.14 |
| chr1_26485721_26485872 | 0.34 |
FAM110D |
family with sequence similarity 110, member D |
285 |
0.83 |
| chr9_131486813_131486981 | 0.34 |
RP11-545E17.3 |
|
173 |
0.76 |
| chr6_32935556_32936303 | 0.34 |
BRD2 |
bromodomain containing 2 |
508 |
0.51 |
| chr10_86559929_86560080 | 0.34 |
ENSG00000238469 |
. |
63017 |
0.16 |
| chr10_104953208_104953414 | 0.33 |
NT5C2 |
5'-nucleotidase, cytosolic II |
255 |
0.93 |
| chr19_50269466_50269673 | 0.33 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
656 |
0.43 |
| chr10_98346955_98347114 | 0.33 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
175 |
0.95 |
| chr17_27942135_27942628 | 0.33 |
CORO6 |
coronin 6 |
2754 |
0.11 |
| chr1_226842452_226842603 | 0.33 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
4844 |
0.24 |
| chr9_115095390_115095852 | 0.32 |
PTBP3 |
polypyrimidine tract binding protein 3 |
237 |
0.93 |
| chr11_10826698_10826955 | 0.32 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
2183 |
0.2 |
| chr19_18209757_18210181 | 0.32 |
IL12RB1 |
interleukin 12 receptor, beta 1 |
215 |
0.88 |
| chr14_22591595_22591746 | 0.32 |
ENSG00000238634 |
. |
19217 |
0.26 |
| chr17_54912094_54912323 | 0.32 |
DGKE |
diacylglycerol kinase, epsilon 64kDa |
119 |
0.92 |
| chr17_58041207_58042016 | 0.31 |
RNFT1 |
ring finger protein, transmembrane 1 |
469 |
0.76 |
| chr4_6200889_6201238 | 0.31 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
1219 |
0.6 |
| chr3_178866443_178867773 | 0.31 |
PIK3CA |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
207 |
0.94 |
| chr17_40729951_40730261 | 0.31 |
PSMC3IP |
PSMC3 interacting protein |
257 |
0.81 |
| chr4_152787665_152787816 | 0.31 |
ENSG00000253077 |
. |
105025 |
0.07 |
| chr6_90061473_90061759 | 0.30 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
951 |
0.59 |
| chr14_89925846_89925997 | 0.30 |
FOXN3 |
forkhead box N3 |
34474 |
0.15 |
| chr12_7995756_7996489 | 0.30 |
ENSG00000222978 |
. |
3762 |
0.15 |
| chr20_47365732_47365883 | 0.30 |
ENSG00000251876 |
. |
9822 |
0.28 |
| chr19_21646219_21646922 | 0.30 |
ZNF429 |
zinc finger protein 429 |
41796 |
0.16 |
| chr7_142161599_142161750 | 0.30 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
172065 |
0.03 |
| chr14_24550083_24550870 | 0.29 |
NRL |
neural retina leucine zipper |
661 |
0.47 |
| chr3_71730862_71731013 | 0.29 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
43589 |
0.15 |
| chr19_59049656_59050353 | 0.29 |
ZBTB45 |
zinc finger and BTB domain containing 45 |
274 |
0.78 |
| chr1_212340351_212340502 | 0.29 |
ENSG00000252879 |
. |
67408 |
0.09 |
| chr3_171825160_171825429 | 0.29 |
FNDC3B |
fibronectin type III domain containing 3B |
19470 |
0.26 |
| chr1_150536035_150536186 | 0.29 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
2141 |
0.14 |
| chr16_89043094_89043288 | 0.29 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
210 |
0.92 |
| chr6_32159535_32159686 | 0.29 |
GPSM3 |
G-protein signaling modulator 3 |
1035 |
0.26 |
| chr17_4336857_4337447 | 0.29 |
SPNS3 |
spinster homolog 3 (Drosophila) |
67 |
0.97 |
| chr20_5584278_5584429 | 0.28 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
7319 |
0.28 |
| chr16_30486681_30486832 | 0.28 |
ITGAL |
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
1434 |
0.16 |
| chr11_134200417_134200568 | 0.28 |
GLB1L2 |
galactosidase, beta 1-like 2 |
1276 |
0.47 |
| chr17_14332143_14332294 | 0.28 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
127818 |
0.06 |
| chr2_106388828_106388979 | 0.28 |
NCK2 |
NCK adaptor protein 2 |
26715 |
0.24 |
| chr19_18606740_18606891 | 0.28 |
ELL |
elongation factor RNA polymerase II |
20072 |
0.09 |
| chr15_65476466_65477266 | 0.28 |
CLPX |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
44 |
0.97 |
| chr13_41576254_41576405 | 0.28 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
17121 |
0.19 |
| chr4_184312949_184313100 | 0.27 |
ENSG00000263395 |
. |
7318 |
0.18 |
| chr11_75063727_75063930 | 0.27 |
ARRB1 |
arrestin, beta 1 |
955 |
0.48 |
| chr5_1501743_1501894 | 0.27 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
22274 |
0.2 |
| chr5_171061425_171061576 | 0.27 |
ENSG00000264303 |
. |
136034 |
0.04 |
| chr22_21024096_21024247 | 0.27 |
ENSG00000264718 |
. |
12849 |
0.14 |
| chr1_28415312_28415525 | 0.27 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
211 |
0.92 |
| chr11_125461662_125462346 | 0.27 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
397 |
0.49 |
| chr1_161185105_161185306 | 0.27 |
FCER1G |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
118 |
0.89 |
| chr7_75543939_75544219 | 0.26 |
POR |
P450 (cytochrome) oxidoreductase |
318 |
0.58 |
| chr6_32144901_32145530 | 0.26 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
113 |
0.87 |
| chr2_240207404_240207640 | 0.26 |
ENSG00000265215 |
. |
19635 |
0.17 |
| chr1_225966293_225966491 | 0.26 |
SRP9 |
signal recognition particle 9kDa |
755 |
0.6 |
| chr18_47003055_47003217 | 0.26 |
RP11-110H1.4 |
|
9383 |
0.08 |
| chr10_105667946_105668135 | 0.26 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
9387 |
0.18 |
| chr9_132647660_132648047 | 0.26 |
FNBP1 |
formin binding protein 1 |
33736 |
0.12 |
| chr6_44042101_44042381 | 0.25 |
RP5-1120P11.1 |
|
148 |
0.95 |
| chr1_233463556_233464041 | 0.25 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
284 |
0.94 |
| chr15_66993020_66993328 | 0.25 |
SMAD6 |
SMAD family member 6 |
1392 |
0.51 |
| chr21_34671674_34671958 | 0.25 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
24918 |
0.13 |
| chr6_126067758_126068417 | 0.25 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
723 |
0.57 |
| chr1_67518540_67519349 | 0.25 |
SLC35D1 |
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
214 |
0.95 |
| chr1_22351357_22351739 | 0.25 |
ENSG00000201273 |
. |
14070 |
0.09 |
| chrX_39953289_39953491 | 0.25 |
BCOR |
BCL6 corepressor |
3266 |
0.39 |
| chr16_30393182_30393494 | 0.25 |
SEPT1 |
septin 1 |
414 |
0.64 |
| chr8_101320634_101321676 | 0.25 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
599 |
0.79 |
| chr12_131246546_131246697 | 0.25 |
RIMBP2 |
RIMS binding protein 2 |
45795 |
0.14 |
| chr17_67322489_67323211 | 0.25 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
381 |
0.89 |
| chr2_216945952_216946469 | 0.25 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
329 |
0.53 |
| chr7_8181769_8181920 | 0.25 |
ENSG00000265212 |
. |
12808 |
0.21 |
| chr1_226593332_226593483 | 0.25 |
PARP1 |
poly (ADP-ribose) polymerase 1 |
2276 |
0.33 |
| chr7_155089675_155090099 | 0.24 |
INSIG1 |
insulin induced gene 1 |
288 |
0.92 |
| chrX_153775545_153775808 | 0.24 |
G6PD |
glucose-6-phosphate dehydrogenase |
96 |
0.49 |
| chr6_143165995_143166218 | 0.24 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
7922 |
0.29 |
| chr7_75796315_75796558 | 0.24 |
AC005077.5 |
|
6615 |
0.18 |
| chr3_71773124_71773375 | 0.24 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
1277 |
0.5 |
| chr1_101749663_101749814 | 0.24 |
RP4-575N6.5 |
|
41024 |
0.13 |
| chr9_95791041_95791204 | 0.24 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
13780 |
0.18 |
| chrX_117631127_117631452 | 0.24 |
DOCK11 |
dedicator of cytokinesis 11 |
1417 |
0.52 |
| chr17_72463483_72463716 | 0.24 |
CD300A |
CD300a molecule |
598 |
0.68 |
| chr12_124873885_124874250 | 0.23 |
NCOR2 |
nuclear receptor corepressor 2 |
697 |
0.79 |
| chr5_77160779_77160930 | 0.23 |
TBCA |
tubulin folding cofactor A |
3750 |
0.36 |
| chr19_17622493_17622725 | 0.23 |
PGLS |
6-phosphogluconolactonase |
65 |
0.94 |
| chr1_149898902_149899634 | 0.23 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
215 |
0.86 |
| chr3_182511875_182512794 | 0.23 |
ATP11B |
ATPase, class VI, type 11B |
1043 |
0.43 |
| chr2_28824878_28825070 | 0.23 |
PLB1 |
phospholipase B1 |
188 |
0.96 |
| chr10_89628293_89628531 | 0.23 |
KLLN |
killin, p53-regulated DNA replication inhibitor |
5218 |
0.15 |
| chr17_74489731_74490033 | 0.23 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
598 |
0.59 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.6 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.6 | GO:0090201 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.1 | 0.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.3 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.3 | GO:0072540 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.2 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.3 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.5 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone methylation(GO:0031060) regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.7 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:1904377 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:0022038 | cerebral cortex radial glia guided migration(GO:0021801) layer formation in cerebral cortex(GO:0021819) telencephalon glial cell migration(GO:0022030) corpus callosum development(GO:0022038) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.2 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.9 | GO:0030330 | DNA damage response, signal transduction by p53 class mediator(GO:0030330) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.5 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |