Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
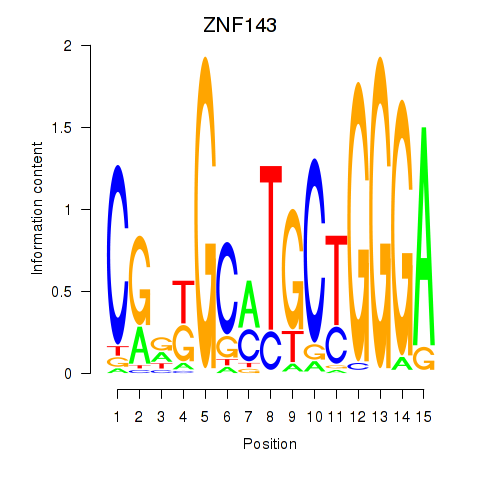
Results for ZNF143
Z-value: 1.50
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.5 | zinc finger protein 143 |
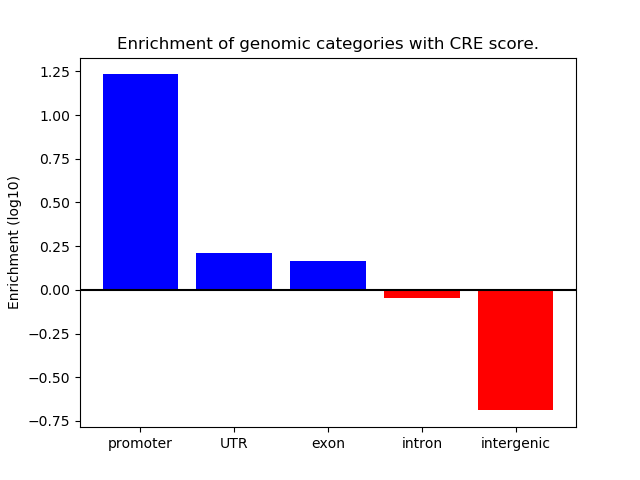
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_9548903_9549060 | ZNF143 | 14937 | 0.157316 | -0.63 | 6.6e-02 | Click! |
| chr11_9510814_9510966 | ZNF143 | 23154 | 0.139994 | -0.55 | 1.3e-01 | Click! |
| chr11_9511154_9511374 | ZNF143 | 22780 | 0.140892 | -0.48 | 2.0e-01 | Click! |
| chr11_9482658_9482912 | ZNF143 | 181 | 0.862863 | 0.43 | 2.5e-01 | Click! |
| chr11_9555045_9555196 | ZNF143 | 21076 | 0.142993 | 0.39 | 3.0e-01 | Click! |
Activity of the ZNF143 motif across conditions
Conditions sorted by the z-value of the ZNF143 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
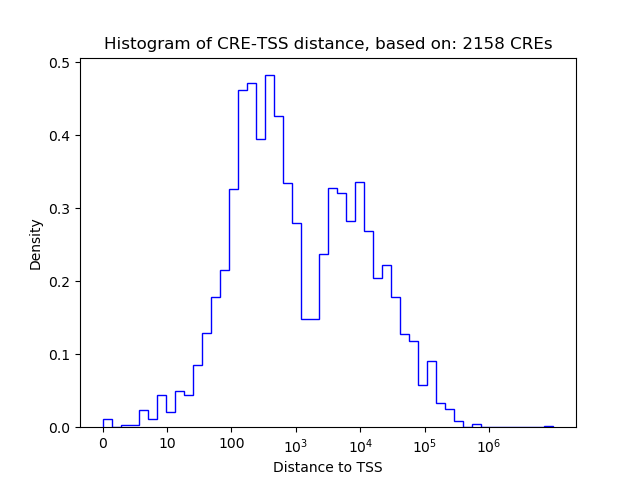
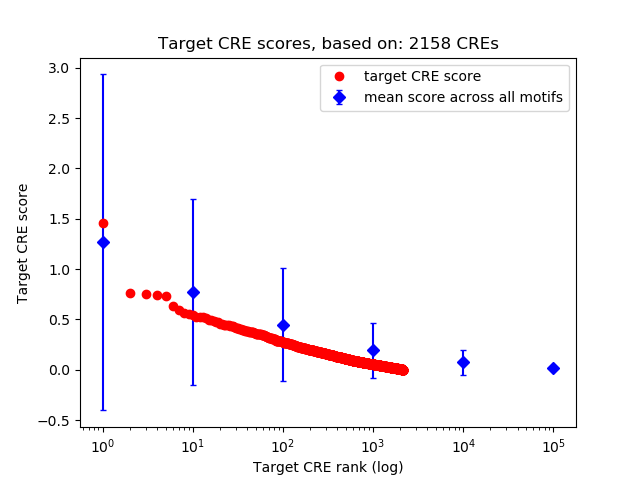
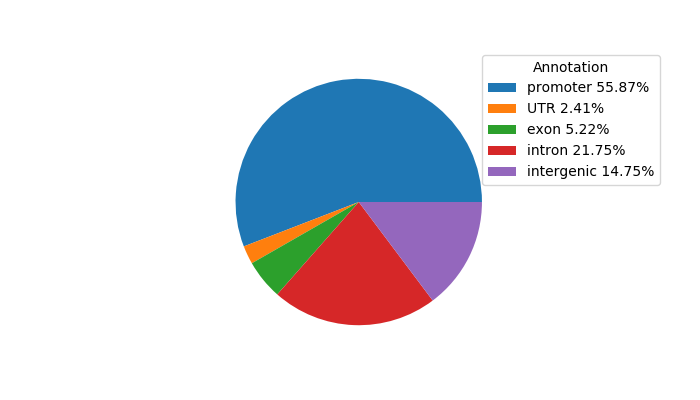
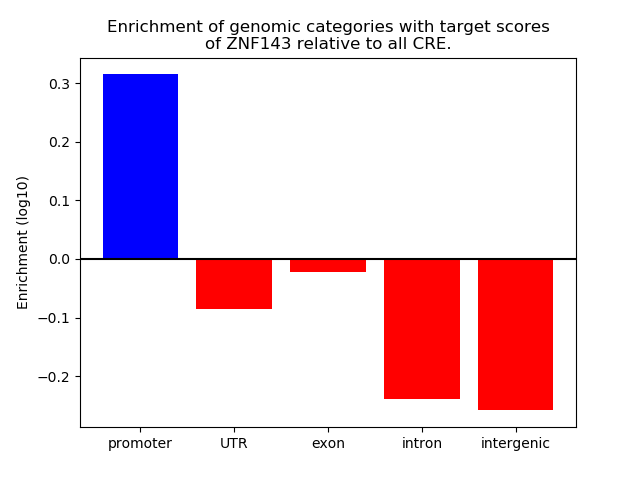
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_135089555_135089795 | 1.46 |
ADAM8 |
ADAM metallopeptidase domain 8 |
679 |
0.52 |
| chr19_48833249_48833485 | 0.76 |
EMP3 |
epithelial membrane protein 3 |
4502 |
0.13 |
| chr2_42599068_42599219 | 0.75 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
2993 |
0.32 |
| chr18_21464195_21464346 | 0.74 |
LAMA3 |
laminin, alpha 3 |
467 |
0.84 |
| chr7_150755256_150755900 | 0.73 |
CDK5 |
cyclin-dependent kinase 5 |
39 |
0.77 |
| chr17_43302789_43303077 | 0.63 |
CTD-2020K17.1 |
|
3344 |
0.12 |
| chrX_152200996_152201209 | 0.59 |
PNMA3 |
paraneoplastic Ma antigen 3 |
23664 |
0.12 |
| chr22_20067197_20067578 | 0.56 |
DGCR8 |
DGCR8 microprocessor complex subunit |
368 |
0.7 |
| chr10_74095282_74095433 | 0.56 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
9362 |
0.19 |
| chr19_17515989_17516450 | 0.54 |
BST2 |
bone marrow stromal cell antigen 2 |
238 |
0.54 |
| chr2_2617005_2617869 | 0.52 |
MYT1L |
myelin transcription factor 1-like |
282471 |
0.01 |
| chr2_74756873_74757049 | 0.52 |
AUP1 |
ancient ubiquitous protein 1 |
105 |
0.55 |
| chr17_56410837_56411117 | 0.52 |
MIR142 |
microRNA 142 |
1108 |
0.32 |
| chr19_10381757_10382103 | 0.51 |
ICAM1 |
intercellular adhesion molecule 1 |
134 |
0.89 |
| chr19_1407342_1407493 | 0.50 |
DAZAP1 |
DAZ associated protein 1 |
151 |
0.88 |
| chr11_2550000_2550151 | 0.49 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
66958 |
0.09 |
| chr8_145734147_145734421 | 0.48 |
MFSD3 |
major facilitator superfamily domain containing 3 |
173 |
0.83 |
| chr12_53894777_53895686 | 0.47 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
150 |
0.79 |
| chr9_95826666_95826817 | 0.47 |
SUSD3 |
sushi domain containing 3 |
4641 |
0.2 |
| chr1_6479504_6479655 | 0.45 |
HES2 |
hes family bHLH transcription factor 2 |
384 |
0.76 |
| chr2_242556723_242557374 | 0.45 |
THAP4 |
THAP domain containing 4 |
132 |
0.94 |
| chr11_809579_809818 | 0.45 |
RPLP2 |
ribosomal protein, large, P2 |
51 |
0.91 |
| chr17_76250693_76250844 | 0.44 |
TMEM235 |
transmembrane protein 235 |
22646 |
0.12 |
| chr2_74669340_74669491 | 0.44 |
RTKN |
rhotekin |
389 |
0.66 |
| chr10_124134109_124134260 | 0.44 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
28 |
0.98 |
| chr2_26100038_26101197 | 0.44 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
718 |
0.76 |
| chr1_939046_939197 | 0.44 |
HES4 |
hes family bHLH transcription factor 4 |
3569 |
0.11 |
| chr7_6199542_6199693 | 0.43 |
CYTH3 |
cytohesin 3 |
13089 |
0.14 |
| chr21_45579276_45579734 | 0.43 |
AP001055.1 |
|
14075 |
0.13 |
| chr19_2562679_2562830 | 0.42 |
CTC-265F19.2 |
|
49106 |
0.09 |
| chrX_49042899_49043187 | 0.41 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
198 |
0.86 |
| chr7_45076135_45076286 | 0.41 |
CCM2 |
cerebral cavernous malformation 2 |
8939 |
0.15 |
| chr14_36295319_36295470 | 0.41 |
BRMS1L |
breast cancer metastasis-suppressor 1-like |
130 |
0.86 |
| chr10_73532392_73533248 | 0.40 |
C10orf54 |
chromosome 10 open reading frame 54 |
435 |
0.83 |
| chr2_178417541_178417760 | 0.40 |
TTC30B |
tetratricopeptide repeat domain 30B |
92 |
0.97 |
| chr6_31802179_31802624 | 0.39 |
C6orf48 |
chromosome 6 open reading frame 48 |
3 |
0.87 |
| chr19_36398415_36398818 | 0.39 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
533 |
0.58 |
| chr2_106362063_106362974 | 0.39 |
NCK2 |
NCK adaptor protein 2 |
330 |
0.94 |
| chr3_69591746_69591897 | 0.38 |
FRMD4B |
FERM domain containing 4B |
87 |
0.99 |
| chr8_141517648_141517821 | 0.38 |
CTA-204B4.2 |
|
408 |
0.85 |
| chr19_55629138_55629447 | 0.38 |
PPP1R12C |
protein phosphatase 1, regulatory subunit 12C |
365 |
0.75 |
| chr16_84582878_84583147 | 0.38 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
4627 |
0.21 |
| chr13_41768167_41768864 | 0.38 |
KBTBD7 |
kelch repeat and BTB (POZ) domain containing 7 |
187 |
0.94 |
| chr5_1004071_1004222 | 0.38 |
ENSG00000221244 |
. |
156 |
0.93 |
| chr10_46168316_46168717 | 0.37 |
ZFAND4 |
zinc finger, AN1-type domain 4 |
288 |
0.92 |
| chr12_110906550_110906725 | 0.37 |
GPN3 |
GPN-loop GTPase 3 |
111 |
0.83 |
| chr17_80260833_80260984 | 0.37 |
CD7 |
CD7 molecule |
14520 |
0.1 |
| chr8_142413039_142413204 | 0.37 |
CTD-3064M3.4 |
|
10447 |
0.12 |
| chr1_26868114_26868336 | 0.36 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
1378 |
0.34 |
| chr3_57038475_57038626 | 0.36 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
27147 |
0.18 |
| chrX_147462854_147463013 | 0.36 |
AC002368.4 |
|
119202 |
0.06 |
| chr19_4791521_4791672 | 0.36 |
FEM1A |
fem-1 homolog a (C. elegans) |
132 |
0.93 |
| chr3_48230084_48230363 | 0.36 |
CDC25A |
cell division cycle 25A |
331 |
0.83 |
| chr1_200008337_200008553 | 0.35 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
3272 |
0.23 |
| chr1_27989345_27989660 | 0.35 |
RP11-288L9.4 |
|
6477 |
0.12 |
| chr19_13274647_13274798 | 0.35 |
CTC-250I14.6 |
|
9715 |
0.11 |
| chr14_81938636_81938919 | 0.35 |
STON2 |
stonin 2 |
35968 |
0.21 |
| chr14_91096638_91096789 | 0.35 |
RP11-1078H9.5 |
|
12269 |
0.18 |
| chr19_2093663_2094074 | 0.35 |
MOB3A |
MOB kinase activator 3A |
2401 |
0.16 |
| chr1_154986335_154986486 | 0.35 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
514 |
0.55 |
| chr8_144634608_144634759 | 0.35 |
GSDMD |
gasdermin D |
694 |
0.41 |
| chr15_41913046_41914152 | 0.34 |
MGA |
MGA, MAX dimerization protein |
135 |
0.96 |
| chr22_45706838_45707043 | 0.34 |
FAM118A |
family with sequence similarity 118, member A |
935 |
0.58 |
| chr1_28974832_28975188 | 0.34 |
ENSG00000270103 |
. |
102 |
0.95 |
| chr3_182879726_182880740 | 0.34 |
LAMP3 |
lysosomal-associated membrane protein 3 |
430 |
0.86 |
| chr7_140098327_140098552 | 0.33 |
SLC37A3 |
solute carrier family 37, member 3 |
90 |
0.96 |
| chr20_5100236_5100597 | 0.33 |
PCNA |
proliferating cell nuclear antigen |
256 |
0.85 |
| chr1_2165353_2165549 | 0.33 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
5317 |
0.14 |
| chr19_48673594_48673912 | 0.32 |
LIG1 |
ligase I, DNA, ATP-dependent |
107 |
0.61 |
| chr19_5255691_5255842 | 0.32 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
30408 |
0.19 |
| chr16_279456_279667 | 0.32 |
LUC7L |
LUC7-like (S. cerevisiae) |
99 |
0.93 |
| chr17_81047529_81047808 | 0.31 |
METRNL |
meteorin, glial cell differentiation regulator-like |
4326 |
0.27 |
| chr3_5028227_5029021 | 0.31 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
6978 |
0.21 |
| chr8_21754477_21754628 | 0.31 |
DOK2 |
docking protein 2, 56kDa |
16622 |
0.18 |
| chr17_28927099_28927365 | 0.31 |
SMURF2P1 |
SMAD specific E3 ubiquitin protein ligase 2 pseudogene 1 |
651 |
0.67 |
| chr22_39637649_39637800 | 0.31 |
PDGFB |
platelet-derived growth factor beta polypeptide |
589 |
0.68 |
| chr11_64005411_64005562 | 0.31 |
VEGFB |
vascular endothelial growth factor B |
478 |
0.51 |
| chr14_39901349_39901640 | 0.31 |
FBXO33 |
F-box protein 33 |
176 |
0.97 |
| chr16_70487949_70488475 | 0.31 |
FUK |
fucokinase |
112 |
0.94 |
| chr19_44331131_44331711 | 0.31 |
ZNF283 |
zinc finger protein 283 |
23 |
0.53 |
| chr19_3801497_3801822 | 0.31 |
MATK |
megakaryocyte-associated tyrosine kinase |
138 |
0.93 |
| chr5_175958496_175958647 | 0.30 |
RNF44 |
ring finger protein 44 |
925 |
0.45 |
| chr20_1446772_1447463 | 0.29 |
NSFL1C |
NSFL1 (p97) cofactor (p47) |
356 |
0.84 |
| chr6_150070946_150071995 | 0.29 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
290 |
0.87 |
| chr19_49004084_49004235 | 0.29 |
LMTK3 |
lemur tyrosine kinase 3 |
10902 |
0.1 |
| chr7_112090576_112091250 | 0.29 |
IFRD1 |
interferon-related developmental regulator 1 |
162 |
0.96 |
| chr6_42332930_42333081 | 0.29 |
ENSG00000221252 |
. |
47129 |
0.14 |
| chr9_131012193_131012363 | 0.29 |
ENSG00000264823 |
. |
4969 |
0.09 |
| chr11_93394564_93394780 | 0.28 |
KIAA1731 |
KIAA1731 |
133 |
0.94 |
| chr19_17517338_17517489 | 0.28 |
MVB12A |
multivesicular body subunit 12A |
459 |
0.54 |
| chr12_6570769_6570920 | 0.28 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
8967 |
0.09 |
| chr19_17634183_17634515 | 0.28 |
FAM129C |
family with sequence similarity 129, member C |
239 |
0.85 |
| chr22_37542366_37542517 | 0.28 |
IL2RB |
interleukin 2 receptor, beta |
3589 |
0.14 |
| chr8_27631821_27631972 | 0.28 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
7 |
0.92 |
| chrX_48814848_48815217 | 0.28 |
OTUD5 |
OTU domain containing 5 |
139 |
0.9 |
| chr4_6675763_6675979 | 0.28 |
AC093323.1 |
Uncharacterized protein |
18318 |
0.14 |
| chr9_130497113_130497635 | 0.28 |
TOR2A |
torsin family 2, member A |
197 |
0.88 |
| chr19_36618863_36619073 | 0.28 |
CAPNS1 |
calpain, small subunit 1 |
11509 |
0.09 |
| chr12_96429136_96429440 | 0.28 |
LTA4H |
leukotriene A4 hydrolase |
151 |
0.95 |
| chr17_62658617_62658986 | 0.28 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
615 |
0.77 |
| chr1_53164097_53164296 | 0.28 |
COA7 |
cytochrome c oxidase assembly factor 7 |
158 |
0.94 |
| chr8_41907411_41908748 | 0.27 |
KAT6A |
K(lysine) acetyltransferase 6A |
1426 |
0.43 |
| chr15_75315646_75315949 | 0.27 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
99 |
0.96 |
| chr16_89042188_89042390 | 0.27 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
1112 |
0.45 |
| chr7_63386955_63387300 | 0.27 |
ENSG00000265017 |
. |
87 |
0.98 |
| chr10_1696776_1696927 | 0.27 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
82819 |
0.11 |
| chr7_72936423_72936636 | 0.27 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
73 |
0.96 |
| chr15_86337104_86338160 | 0.27 |
KLHL25 |
kelch-like family member 25 |
468 |
0.73 |
| chr1_181451777_181451943 | 0.27 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
856 |
0.76 |
| chrX_118823023_118823250 | 0.27 |
SEPT6 |
septin 6 |
3656 |
0.22 |
| chr7_72439858_72440009 | 0.27 |
TRIM74 |
tripartite motif containing 74 |
64 |
0.95 |
| chr16_88366626_88367008 | 0.26 |
ZNF469 |
zinc finger protein 469 |
127062 |
0.05 |
| chr10_79613508_79613659 | 0.26 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
13050 |
0.18 |
| chr9_38621823_38622122 | 0.26 |
FAM201A |
family with sequence similarity 201, member A |
887 |
0.5 |
| chr1_117594372_117594685 | 0.26 |
RP11-27K13.3 |
|
7584 |
0.17 |
| chr17_8076861_8077643 | 0.26 |
ENSG00000200463 |
. |
347 |
0.7 |
| chr17_26874549_26874726 | 0.26 |
UNC119 |
unc-119 homolog (C. elegans) |
1780 |
0.17 |
| chr1_16470291_16470442 | 0.26 |
RP11-276H7.2 |
|
11340 |
0.12 |
| chr2_240187102_240187645 | 0.26 |
ENSG00000265215 |
. |
39784 |
0.13 |
| chr7_26191457_26191700 | 0.26 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
282 |
0.91 |
| chr15_94406450_94406737 | 0.26 |
ENSG00000222409 |
. |
33663 |
0.25 |
| chr19_39811900_39812051 | 0.25 |
CTC-246B18.8 |
|
477 |
0.64 |
| chr18_23670660_23670942 | 0.25 |
SS18 |
synovial sarcoma translocation, chromosome 18 |
64 |
0.98 |
| chr19_45393848_45394047 | 0.25 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
121 |
0.58 |
| chr14_103801113_103802028 | 0.25 |
EIF5 |
eukaryotic translation initiation factor 5 |
284 |
0.87 |
| chr2_96782390_96782846 | 0.25 |
ADRA2B |
adrenoceptor alpha 2B |
634 |
0.68 |
| chr5_176944022_176944323 | 0.25 |
DDX41 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
255 |
0.85 |
| chr1_32645201_32645477 | 0.25 |
TXLNA |
taxilin alpha |
52 |
0.94 |
| chr7_134670907_134671205 | 0.25 |
AGBL3 |
ATP/GTP binding protein-like 3 |
203 |
0.96 |
| chr17_9330961_9331112 | 0.25 |
AC087501.1 |
|
24478 |
0.18 |
| chr14_75926184_75926335 | 0.25 |
JDP2 |
Jun dimerization protein 2 |
27422 |
0.16 |
| chr12_56498056_56498239 | 0.25 |
PA2G4 |
proliferation-associated 2G4, 38kDa |
44 |
0.92 |
| chr19_49972030_49972206 | 0.25 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
5367 |
0.06 |
| chr11_112097343_112097611 | 0.24 |
PTS |
6-pyruvoyltetrahydropterin synthase |
322 |
0.82 |
| chr22_37937663_37937814 | 0.24 |
RP5-1177I5.3 |
|
8295 |
0.12 |
| chr11_119562641_119562792 | 0.24 |
ENSG00000199217 |
. |
35695 |
0.14 |
| chrX_132091720_132091956 | 0.24 |
HS6ST2 |
heparan sulfate 6-O-sulfotransferase 2 |
494 |
0.87 |
| chr1_151226917_151227146 | 0.24 |
PSMD4 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
148 |
0.91 |
| chr22_42696607_42696758 | 0.24 |
TCF20 |
transcription factor 20 (AR1) |
42940 |
0.14 |
| chr1_28195302_28195741 | 0.24 |
THEMIS2 |
thymocyte selection associated family member 2 |
3534 |
0.14 |
| chr14_91868738_91869034 | 0.23 |
CCDC88C |
coiled-coil domain containing 88C |
14804 |
0.23 |
| chr1_207243797_207243948 | 0.23 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
5523 |
0.14 |
| chr3_9773477_9774131 | 0.23 |
BRPF1 |
bromodomain and PHD finger containing, 1 |
332 |
0.81 |
| chr12_89747023_89747227 | 0.23 |
DUSP6 |
dual specificity phosphatase 6 |
77 |
0.98 |
| chr9_95794200_95794351 | 0.23 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
16933 |
0.17 |
| chr3_158288936_158289364 | 0.23 |
MLF1 |
myeloid leukemia factor 1 |
100 |
0.76 |
| chr19_13952735_13952973 | 0.23 |
ENSG00000207980 |
. |
5381 |
0.08 |
| chr1_1689799_1689950 | 0.23 |
NADK |
NAD kinase |
140 |
0.94 |
| chr21_17959283_17959568 | 0.23 |
ENSG00000207863 |
. |
3132 |
0.31 |
| chr4_40058976_40059529 | 0.23 |
N4BP2 |
NEDD4 binding protein 2 |
696 |
0.68 |
| chr9_139581476_139581762 | 0.23 |
AGPAT2 |
1-acylglycerol-3-phosphate O-acyltransferase 2 |
229 |
0.84 |
| chr18_3447400_3448025 | 0.23 |
TGIF1 |
TGFB-induced factor homeobox 1 |
105 |
0.97 |
| chr15_72522652_72522803 | 0.23 |
PKM |
pyruvate kinase, muscle |
664 |
0.65 |
| chr22_37618634_37618785 | 0.23 |
SSTR3 |
somatostatin receptor 3 |
10347 |
0.13 |
| chr3_150126096_150126425 | 0.23 |
TSC22D2 |
TSC22 domain family, member 2 |
138 |
0.98 |
| chr19_1260409_1260809 | 0.22 |
CIRBP |
cold inducible RNA binding protein |
546 |
0.56 |
| chr5_142177952_142178103 | 0.22 |
ARHGAP26 |
Rho GTPase activating protein 26 |
25745 |
0.22 |
| chr6_36164778_36164969 | 0.22 |
RP1-179N16.6 |
|
107 |
0.53 |
| chr19_45257252_45257704 | 0.22 |
BCL3 |
B-cell CLL/lymphoma 3 |
2900 |
0.15 |
| chr15_90447970_90448121 | 0.22 |
C15orf38 |
chromosome 15 open reading frame 38 |
870 |
0.42 |
| chr7_65579625_65579846 | 0.22 |
CRCP |
CGRP receptor component |
78 |
0.97 |
| chr4_6687752_6687903 | 0.22 |
AC093323.1 |
Uncharacterized protein |
6362 |
0.15 |
| chrX_65234090_65234241 | 0.22 |
ENSG00000207939 |
. |
4547 |
0.23 |
| chr11_73358513_73358664 | 0.22 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
6 |
0.98 |
| chr11_18720549_18721118 | 0.22 |
TMEM86A |
transmembrane protein 86A |
495 |
0.74 |
| chr2_223169880_223170456 | 0.22 |
CCDC140 |
coiled-coil domain containing 140 |
2471 |
0.27 |
| chr4_1581349_1582263 | 0.22 |
FAM53A |
family with sequence similarity 53, member A |
75329 |
0.08 |
| chr16_21170219_21170819 | 0.22 |
DNAH3 |
dynein, axonemal, heavy chain 3 |
243 |
0.71 |
| chr6_159464305_159464462 | 0.22 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1667 |
0.38 |
| chr7_75024883_75025034 | 0.22 |
TRIM73 |
tripartite motif containing 73 |
55 |
0.84 |
| chr1_6532288_6532439 | 0.22 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
6108 |
0.12 |
| chr17_40440128_40440459 | 0.22 |
STAT5A |
signal transducer and activator of transcription 5A |
194 |
0.91 |
| chr5_131831286_131831908 | 0.21 |
IRF1 |
interferon regulatory factor 1 |
5107 |
0.15 |
| chr1_231917497_231917650 | 0.21 |
ENSG00000222986 |
. |
111192 |
0.06 |
| chr1_27248269_27248533 | 0.21 |
NUDC |
nudC nuclear distribution protein |
184 |
0.91 |
| chr2_69002223_69002932 | 0.21 |
ARHGAP25 |
Rho GTPase activating protein 25 |
505 |
0.84 |
| chr19_40596081_40597156 | 0.21 |
AC005614.3 |
|
43 |
0.64 |
| chr9_99539749_99540719 | 0.21 |
ZNF510 |
zinc finger protein 510 |
94 |
0.98 |
| chr18_9673690_9673841 | 0.21 |
RAB31 |
RAB31, member RAS oncogene family |
34397 |
0.15 |
| chr16_31276973_31277152 | 0.21 |
ENSG00000252876 |
. |
1750 |
0.22 |
| chr8_27183780_27184189 | 0.21 |
PTK2B |
protein tyrosine kinase 2 beta |
356 |
0.89 |
| chr1_234861269_234861851 | 0.21 |
ENSG00000201638 |
. |
112160 |
0.06 |
| chr7_26332124_26332363 | 0.21 |
SNX10 |
sorting nexin 10 |
431 |
0.87 |
| chr7_140396423_140396944 | 0.21 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
105 |
0.63 |
| chr1_161592822_161592973 | 0.20 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
7925 |
0.12 |
| chr14_89258910_89259265 | 0.20 |
EML5 |
echinoderm microtubule associated protein like 5 |
9 |
0.98 |
| chr3_71956368_71956519 | 0.20 |
ENSG00000239250 |
. |
80115 |
0.1 |
| chr3_123304144_123304696 | 0.20 |
MYLK-AS1 |
MYLK antisense RNA 1 |
17 |
0.87 |
| chr13_28542515_28542921 | 0.20 |
CDX2 |
caudal type homeobox 2 |
2558 |
0.21 |
| chr3_32858150_32858717 | 0.20 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
1077 |
0.61 |
| chr1_149764655_149765085 | 0.20 |
RP11-196G18.3 |
|
294 |
0.77 |
| chr9_86755575_86756256 | 0.20 |
RP11-380F14.2 |
|
137219 |
0.04 |
| chr11_133797734_133798200 | 0.20 |
IGSF9B |
immunoglobulin superfamily, member 9B |
18220 |
0.18 |
| chr17_36859934_36860212 | 0.20 |
ENSG00000265930 |
. |
1489 |
0.16 |
| chr11_66106791_66106942 | 0.20 |
RIN1 |
Ras and Rab interactor 1 |
2555 |
0.09 |
| chr19_37957537_37958312 | 0.20 |
ZNF569 |
zinc finger protein 569 |
186 |
0.85 |
| chr10_74113543_74114362 | 0.20 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
186 |
0.95 |
| chr1_150540056_150540997 | 0.20 |
ENSG00000264508 |
. |
1190 |
0.22 |
| chr3_51976372_51976799 | 0.20 |
PARP3 |
poly (ADP-ribose) polymerase family, member 3 |
210 |
0.75 |
| chr17_35216667_35216818 | 0.20 |
RP11-445F12.1 |
|
77179 |
0.07 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.2 | 0.6 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.3 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 | 0.4 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0072224 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.1 | 0.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.2 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.2 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.3 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0003136 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.2 | GO:0032647 | interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.0 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.2 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.2 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.0 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.3 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.6 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |