Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
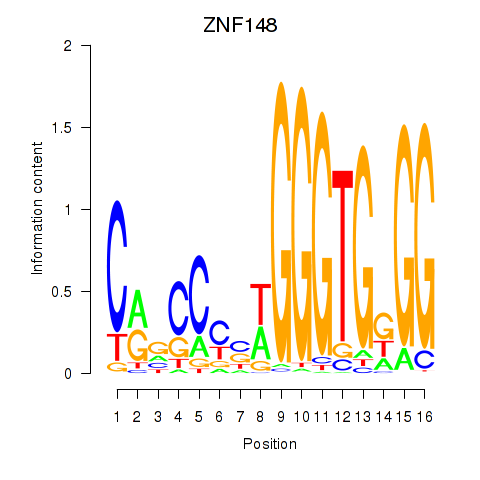
Results for ZNF148
Z-value: 0.94
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | zinc finger protein 148 |
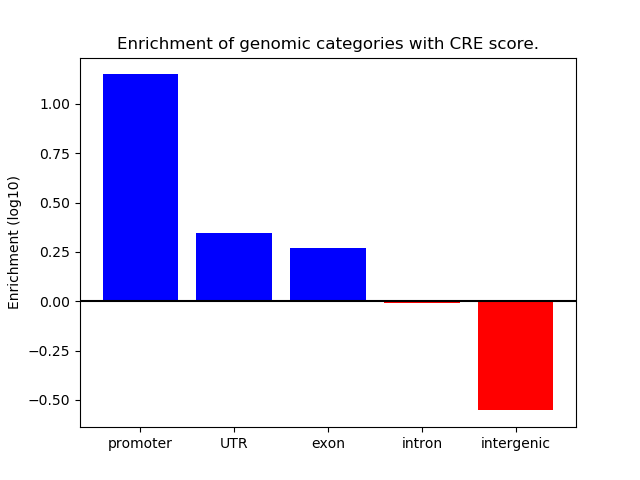
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_125092923_125093609 | ZNF148 | 827 | 0.637282 | -0.63 | 6.9e-02 | Click! |
| chr3_125086590_125086789 | ZNF148 | 7404 | 0.207139 | 0.45 | 2.2e-01 | Click! |
| chr3_125092460_125092611 | ZNF148 | 1558 | 0.404423 | -0.36 | 3.4e-01 | Click! |
| chr3_125076195_125076694 | ZNF148 | 218 | 0.943425 | -0.33 | 3.8e-01 | Click! |
| chr3_125094287_125094942 | ZNF148 | 416 | 0.850436 | 0.31 | 4.1e-01 | Click! |
Activity of the ZNF148 motif across conditions
Conditions sorted by the z-value of the ZNF148 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
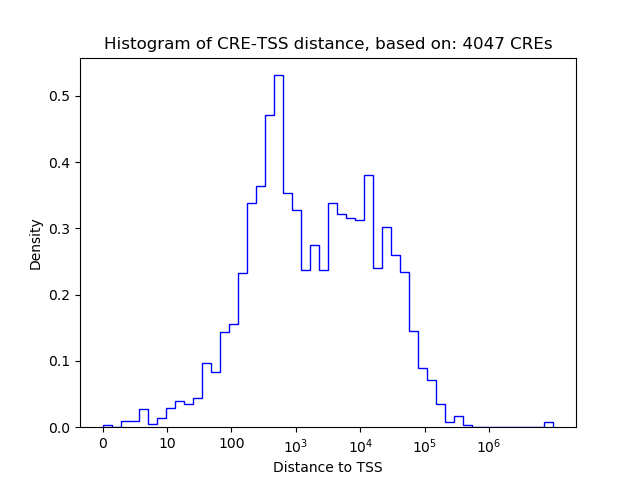
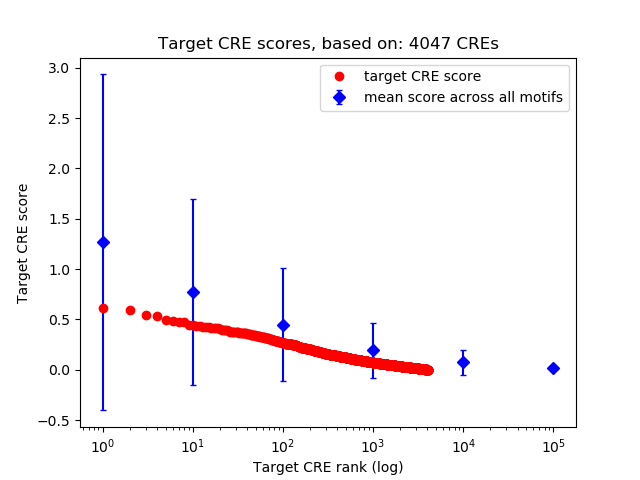
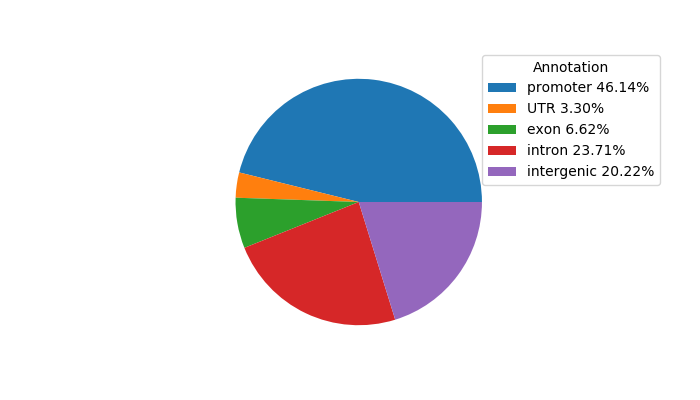
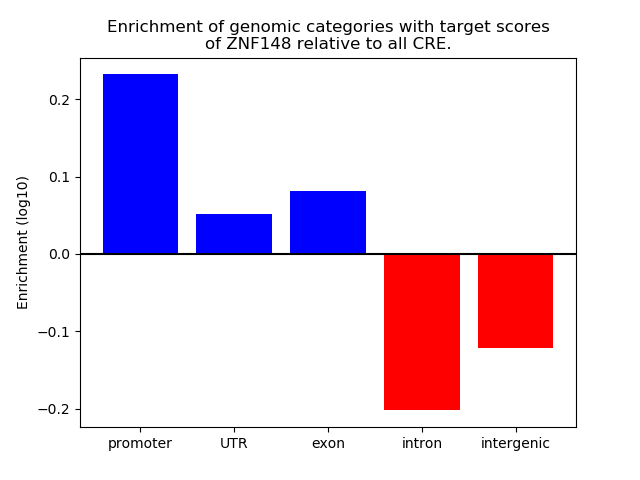
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_8698839_8699470 | 0.61 |
KAL1 |
Kallmann syndrome 1 sequence |
1073 |
0.66 |
| chr10_118031771_118032060 | 0.59 |
GFRA1 |
GDNF family receptor alpha 1 |
121 |
0.98 |
| chr12_48109642_48109793 | 0.55 |
ENDOU |
endonuclease, polyU-specific |
1715 |
0.32 |
| chr11_131780761_131781452 | 0.53 |
NTM |
neurotrimin |
209 |
0.95 |
| chr12_117799295_117799446 | 0.49 |
NOS1 |
nitric oxide synthase 1 (neuronal) |
212 |
0.96 |
| chr18_6729547_6730123 | 0.48 |
ARHGAP28 |
Rho GTPase activating protein 28 |
14 |
0.5 |
| chr19_11253525_11253809 | 0.48 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
12770 |
0.13 |
| chr14_105830576_105830747 | 0.47 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
18795 |
0.13 |
| chr17_73718374_73718525 | 0.45 |
ITGB4 |
integrin, beta 4 |
872 |
0.43 |
| chr4_74809740_74810515 | 0.44 |
PF4 |
platelet factor 4 |
37714 |
0.11 |
| chr6_10383738_10384217 | 0.44 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
20850 |
0.18 |
| chr1_3390049_3390200 | 0.43 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
1936 |
0.35 |
| chr2_9347772_9348251 | 0.43 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
1117 |
0.65 |
| chr6_125684588_125685144 | 0.43 |
RP11-735G4.1 |
|
10604 |
0.27 |
| chr10_126029940_126030091 | 0.42 |
OAT |
ornithine aminotransferase |
77490 |
0.1 |
| chr5_71403321_71403664 | 0.42 |
MAP1B |
microtubule-associated protein 1B |
179 |
0.97 |
| chr6_3455456_3455921 | 0.42 |
SLC22A23 |
solute carrier family 22, member 23 |
589 |
0.84 |
| chr17_3848324_3848628 | 0.41 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
14811 |
0.16 |
| chr10_118764443_118765166 | 0.41 |
KIAA1598 |
KIAA1598 |
72 |
0.98 |
| chr1_27336321_27336472 | 0.40 |
FAM46B |
family with sequence similarity 46, member B |
2931 |
0.22 |
| chr14_105116453_105116770 | 0.40 |
ENSG00000265291 |
. |
27475 |
0.12 |
| chr10_133850335_133850615 | 0.39 |
BNIP3 |
BCL2/adenovirus E1B 19kDa interacting protein 3 |
55040 |
0.13 |
| chr6_43252045_43252196 | 0.39 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
13616 |
0.13 |
| chr1_10891240_10891391 | 0.39 |
CASZ1 |
castor zinc finger 1 |
34608 |
0.18 |
| chr11_65666484_65666730 | 0.38 |
FOSL1 |
FOS-like antigen 1 |
1283 |
0.23 |
| chr4_53729260_53729411 | 0.38 |
RASL11B |
RAS-like, family 11, member B |
878 |
0.66 |
| chr14_104007555_104007706 | 0.37 |
ENSG00000252469 |
. |
9052 |
0.08 |
| chr19_49652849_49653066 | 0.37 |
PPFIA3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
1599 |
0.2 |
| chr2_167232459_167233133 | 0.37 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
293 |
0.93 |
| chr16_15236302_15236453 | 0.37 |
ENSG00000207294 |
. |
8177 |
0.11 |
| chr12_7076786_7076937 | 0.37 |
ENSG00000238795 |
. |
92 |
0.69 |
| chr5_141258225_141258866 | 0.37 |
PCDH1 |
protocadherin 1 |
266 |
0.92 |
| chr8_143224078_143224229 | 0.37 |
ENSG00000265247 |
. |
33547 |
0.19 |
| chr19_33884880_33885031 | 0.37 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
19737 |
0.19 |
| chr9_124360553_124361088 | 0.37 |
RP11-524G24.2 |
|
22327 |
0.18 |
| chr1_17274725_17274876 | 0.37 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
25550 |
0.12 |
| chr16_2174847_2175247 | 0.36 |
RP11-304L19.2 |
|
5839 |
0.06 |
| chr8_72459619_72459990 | 0.36 |
RP11-1102P16.1 |
Uncharacterized protein |
88 |
0.98 |
| chr13_52165897_52166092 | 0.36 |
ENSG00000242893 |
. |
2357 |
0.21 |
| chr2_86055147_86055298 | 0.36 |
ENSG00000252321 |
. |
7487 |
0.17 |
| chr16_4373749_4373941 | 0.36 |
GLIS2 |
GLIS family zinc finger 2 |
8380 |
0.13 |
| chr17_46103279_46103479 | 0.36 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
7859 |
0.09 |
| chr19_17225078_17225229 | 0.35 |
MYO9B |
myosin IXB |
12625 |
0.11 |
| chr16_54321004_54321411 | 0.35 |
IRX3 |
iroquois homeobox 3 |
532 |
0.83 |
| chr10_120739922_120740073 | 0.35 |
RP11-498J9.2 |
|
23399 |
0.15 |
| chr8_144501585_144501736 | 0.35 |
RP11-909N17.3 |
|
1811 |
0.21 |
| chr3_13869046_13869197 | 0.34 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
52497 |
0.13 |
| chr3_139654401_139654885 | 0.34 |
CLSTN2 |
calsyntenin 2 |
616 |
0.84 |
| chr10_35102956_35103650 | 0.34 |
PARD3 |
par-3 family cell polarity regulator |
946 |
0.5 |
| chr1_156890393_156890693 | 0.34 |
LRRC71 |
leucine rich repeat containing 71 |
101 |
0.95 |
| chr7_139899946_139900413 | 0.34 |
ENSG00000199971 |
. |
9292 |
0.16 |
| chr5_6767017_6767234 | 0.34 |
PAPD7 |
PAP associated domain containing 7 |
52380 |
0.15 |
| chr12_49689580_49689731 | 0.34 |
PRPH |
peripherin |
510 |
0.66 |
| chr6_155586253_155586446 | 0.33 |
CLDN20 |
claudin 20 |
1202 |
0.49 |
| chr17_43205399_43205692 | 0.33 |
PLCD3 |
phospholipase C, delta 3 |
4346 |
0.12 |
| chr7_124404967_124405458 | 0.33 |
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
469 |
0.88 |
| chr8_144653425_144654110 | 0.33 |
MROH6 |
maestro heat-like repeat family member 6 |
1151 |
0.21 |
| chr2_160918347_160919586 | 0.33 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
155 |
0.98 |
| chr2_242504094_242504245 | 0.33 |
BOK-AS1 |
BOK antisense RNA 1 |
5777 |
0.13 |
| chr17_16977527_16977700 | 0.33 |
MPRIP-AS1 |
MPRIP antisense RNA 1 |
3453 |
0.23 |
| chr4_1798938_1799089 | 0.33 |
FGFR3 |
fibroblast growth factor receptor 3 |
3390 |
0.22 |
| chr6_150246697_150247451 | 0.32 |
RAET1G |
retinoic acid early transcript 1G |
2817 |
0.17 |
| chr19_33784339_33784950 | 0.32 |
CTD-2540B15.11 |
|
6196 |
0.14 |
| chr11_45202678_45203014 | 0.32 |
PRDM11 |
PR domain containing 11 |
582 |
0.81 |
| chr17_15168005_15168252 | 0.31 |
PMP22 |
peripheral myelin protein 22 |
496 |
0.74 |
| chr21_44774072_44774359 | 0.31 |
SIK1 |
salt-inducible kinase 1 |
72793 |
0.11 |
| chr14_69015254_69015531 | 0.31 |
CTD-2325P2.4 |
|
79770 |
0.1 |
| chr22_46287338_46287489 | 0.31 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
81182 |
0.07 |
| chr7_100464814_100466140 | 0.31 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr3_65779704_65779855 | 0.31 |
MAGI1-AS1 |
MAGI1 antisense RNA 1 |
99712 |
0.08 |
| chr10_128594353_128594780 | 0.31 |
DOCK1 |
dedicator of cytokinesis 1 |
588 |
0.8 |
| chr8_61936210_61936526 | 0.31 |
CLVS1 |
clavesin 1 |
33349 |
0.24 |
| chr5_68710635_68711790 | 0.31 |
MARVELD2 |
MARVEL domain containing 2 |
3 |
0.97 |
| chr15_31734664_31735087 | 0.30 |
KLF13 |
Kruppel-like factor 13 |
76518 |
0.12 |
| chr16_89895218_89895373 | 0.30 |
SPIRE2 |
spire-type actin nucleation factor 2 |
340 |
0.84 |
| chr20_60892287_60892438 | 0.30 |
RP11-157P1.4 |
|
10981 |
0.12 |
| chr11_118489826_118490091 | 0.29 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
11600 |
0.11 |
| chr12_122235363_122235779 | 0.29 |
RHOF |
ras homolog family member F (in filopodia) |
3012 |
0.21 |
| chr16_4369635_4369919 | 0.29 |
GLIS2 |
GLIS family zinc finger 2 |
5015 |
0.15 |
| chr2_131594534_131594685 | 0.29 |
AC133785.1 |
|
42 |
0.87 |
| chr7_1577374_1577649 | 0.29 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
833 |
0.54 |
| chrX_114795286_114795437 | 0.29 |
PLS3 |
plastin 3 |
148 |
0.94 |
| chr16_12995514_12995665 | 0.29 |
SHISA9 |
shisa family member 9 |
25 |
0.98 |
| chr1_155102531_155102783 | 0.29 |
EFNA1 |
ephrin-A1 |
2305 |
0.11 |
| chr8_22723308_22723535 | 0.29 |
RP11-87E22.2 |
|
12064 |
0.17 |
| chr14_71355414_71355574 | 0.29 |
PCNX |
pecanex homolog (Drosophila) |
18628 |
0.26 |
| chr16_85253495_85253759 | 0.29 |
CTC-786C10.1 |
|
48745 |
0.15 |
| chr1_1356913_1357064 | 0.28 |
ANKRD65 |
ankyrin repeat domain 65 |
161 |
0.85 |
| chr1_956379_957022 | 0.28 |
AGRN |
agrin |
1197 |
0.27 |
| chr17_46131994_46132145 | 0.28 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
3 |
0.95 |
| chr19_675778_676178 | 0.28 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
414 |
0.71 |
| chr11_12108503_12108884 | 0.28 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
6850 |
0.29 |
| chr7_100660155_100661047 | 0.28 |
RP11-395B7.4 |
|
281 |
0.85 |
| chr15_90605491_90605664 | 0.28 |
ZNF710 |
zinc finger protein 710 |
5669 |
0.16 |
| chr17_39676556_39676707 | 0.27 |
KRT15 |
keratin 15 |
640 |
0.5 |
| chr7_45002030_45002849 | 0.27 |
RP4-647J21.1 |
|
1931 |
0.25 |
| chr4_715649_715800 | 0.27 |
RP11-1191J2.4 |
|
2099 |
0.18 |
| chr19_38494188_38494741 | 0.27 |
CTC-244M17.1 |
|
39971 |
0.13 |
| chr14_31343280_31344466 | 0.27 |
COCH |
cochlin |
78 |
0.97 |
| chr2_95742439_95743056 | 0.27 |
AC103563.9 |
|
23826 |
0.15 |
| chr22_50740529_50740680 | 0.27 |
PLXNB2 |
plexin B2 |
713 |
0.47 |
| chr1_92495646_92496322 | 0.27 |
EPHX4 |
epoxide hydrolase 4 |
445 |
0.85 |
| chr1_19250411_19250700 | 0.27 |
IFFO2 |
intermediate filament family orphan 2 |
12069 |
0.16 |
| chr1_16343782_16344178 | 0.27 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
340 |
0.8 |
| chr1_109969825_109970028 | 0.26 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
864 |
0.52 |
| chr9_140111635_140112002 | 0.26 |
RNF208 |
ring finger protein 208 |
3957 |
0.07 |
| chr12_125002282_125002834 | 0.26 |
NCOR2 |
nuclear receptor corepressor 2 |
282 |
0.95 |
| chr20_48606144_48606295 | 0.26 |
SNAI1 |
snail family zinc finger 1 |
6683 |
0.17 |
| chr17_35207376_35207682 | 0.26 |
RP11-445F12.1 |
|
86392 |
0.07 |
| chr6_49518031_49518578 | 0.26 |
C6orf141 |
chromosome 6 open reading frame 141 |
191 |
0.95 |
| chr6_43704451_43704602 | 0.26 |
VEGFA |
vascular endothelial growth factor A |
33424 |
0.12 |
| chr9_130318193_130318378 | 0.26 |
FAM129B |
family with sequence similarity 129, member B |
13082 |
0.17 |
| chr7_70079490_70079756 | 0.26 |
AUTS2 |
autism susceptibility candidate 2 |
114502 |
0.07 |
| chr2_157176521_157176855 | 0.26 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
9945 |
0.27 |
| chr21_40393056_40393282 | 0.26 |
ENSG00000272015 |
. |
126460 |
0.05 |
| chr5_131594297_131594770 | 0.26 |
PDLIM4 |
PDZ and LIM domain 4 |
1133 |
0.45 |
| chr1_201228727_201229116 | 0.26 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
23659 |
0.15 |
| chr19_3097024_3097175 | 0.26 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
2691 |
0.16 |
| chr6_36684370_36684521 | 0.26 |
RAB44 |
RAB44, member RAS oncogene family |
1189 |
0.44 |
| chr22_38291029_38291180 | 0.26 |
ENSG00000207227 |
. |
3750 |
0.12 |
| chr19_1748372_1748523 | 0.25 |
ONECUT3 |
one cut homeobox 3 |
3925 |
0.16 |
| chr1_1369498_1369649 | 0.25 |
VWA1 |
von Willebrand factor A domain containing 1 |
668 |
0.44 |
| chr1_156092284_156092708 | 0.25 |
LMNA |
lamin A/C |
3455 |
0.14 |
| chr1_17914943_17915229 | 0.25 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
175 |
0.97 |
| chr19_1789718_1789869 | 0.25 |
ATP8B3 |
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
21830 |
0.09 |
| chr18_77218683_77219016 | 0.25 |
AC018445.1 |
Uncharacterized protein |
57208 |
0.13 |
| chr1_27425556_27426394 | 0.25 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
54998 |
0.1 |
| chr20_36915633_36915784 | 0.25 |
BPI |
bactericidal/permeability-increasing protein |
16817 |
0.15 |
| chr16_68772325_68772476 | 0.25 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
1145 |
0.43 |
| chr6_150070946_150071995 | 0.25 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
290 |
0.87 |
| chr8_27491561_27492240 | 0.25 |
SCARA3 |
scavenger receptor class A, member 3 |
202 |
0.93 |
| chr7_123672325_123672477 | 0.25 |
RP5-921G16.1 |
|
142 |
0.93 |
| chr12_49942443_49943296 | 0.25 |
KCNH3 |
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
9929 |
0.13 |
| chr17_836473_836730 | 0.25 |
RP11-676J12.7 |
Uncharacterized protein |
23729 |
0.14 |
| chr17_75283624_75283888 | 0.25 |
SEPT9 |
septin 9 |
217 |
0.95 |
| chr6_153303042_153303631 | 0.25 |
FBXO5 |
F-box protein 5 |
817 |
0.52 |
| chr1_53951416_53951567 | 0.25 |
DMRTB1 |
DMRT-like family B with proline-rich C-terminal, 1 |
26419 |
0.21 |
| chr9_124527262_124527413 | 0.24 |
DAB2IP |
DAB2 interacting protein |
22273 |
0.22 |
| chr22_50312384_50313103 | 0.24 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
362 |
0.66 |
| chr1_208374584_208374735 | 0.24 |
PLXNA2 |
plexin A2 |
43006 |
0.22 |
| chr14_105436556_105437076 | 0.24 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7878 |
0.16 |
| chr11_707284_707470 | 0.24 |
EPS8L2 |
EPS8-like 2 |
748 |
0.42 |
| chr17_20746972_20747807 | 0.24 |
RP11-344E13.4 |
|
11870 |
0.17 |
| chr20_822913_823164 | 0.24 |
FAM110A |
family with sequence similarity 110, member A |
2247 |
0.36 |
| chr19_2461781_2462465 | 0.24 |
LMNB2 |
lamin B2 |
5129 |
0.15 |
| chr10_79396627_79397649 | 0.24 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
67 |
0.98 |
| chr19_57350948_57351186 | 0.23 |
PEG3 |
paternally expressed 3 |
997 |
0.38 |
| chr1_153536795_153537072 | 0.23 |
S100A2 |
S100 calcium binding protein A2 |
224 |
0.84 |
| chr3_131079896_131080531 | 0.23 |
ENSG00000250608 |
. |
242 |
0.76 |
| chr9_22006120_22006693 | 0.23 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
2546 |
0.22 |
| chr3_30140484_30140635 | 0.23 |
ENSG00000264178 |
. |
39180 |
0.18 |
| chr2_159825200_159826543 | 0.23 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
688 |
0.73 |
| chr2_208370253_208370617 | 0.23 |
CREB1 |
cAMP responsive element binding protein 1 |
24026 |
0.19 |
| chr2_113882342_113882741 | 0.23 |
IL1RN |
interleukin 1 receptor antagonist |
2597 |
0.21 |
| chr4_2935171_2936035 | 0.23 |
MFSD10 |
major facilitator superfamily domain containing 10 |
71 |
0.94 |
| chr1_32166523_32166715 | 0.22 |
COL16A1 |
collagen, type XVI, alpha 1 |
3149 |
0.19 |
| chr19_13112715_13112989 | 0.22 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
6200 |
0.09 |
| chr11_72451789_72452038 | 0.22 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11535 |
0.12 |
| chr1_159842521_159842672 | 0.22 |
RP11-190A12.8 |
|
5852 |
0.1 |
| chr12_102454924_102455862 | 0.22 |
CCDC53 |
coiled-coil domain containing 53 |
464 |
0.76 |
| chr1_225117342_225117684 | 0.22 |
DNAH14 |
dynein, axonemal, heavy chain 14 |
129 |
0.98 |
| chr12_3069053_3070004 | 0.22 |
TEAD4 |
TEA domain family member 4 |
445 |
0.77 |
| chr17_80288187_80288338 | 0.22 |
SECTM1 |
secreted and transmembrane 1 |
1880 |
0.21 |
| chr1_43201434_43201585 | 0.22 |
CLDN19 |
claudin 19 |
4302 |
0.16 |
| chr5_175828477_175828628 | 0.22 |
NOP16 |
NOP16 nucleolar protein |
12576 |
0.1 |
| chr16_88877375_88878305 | 0.22 |
APRT |
adenine phosphoribosyltransferase |
465 |
0.66 |
| chr15_74724718_74725429 | 0.22 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
928 |
0.5 |
| chr5_38845474_38846010 | 0.22 |
OSMR |
oncostatin M receptor |
218 |
0.96 |
| chr1_38510289_38510696 | 0.22 |
POU3F1 |
POU class 3 homeobox 1 |
1958 |
0.27 |
| chr13_33858654_33859881 | 0.22 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
625 |
0.76 |
| chr8_23204118_23204478 | 0.22 |
ENSG00000253837 |
. |
10577 |
0.16 |
| chr9_34578314_34578465 | 0.22 |
CNTFR-AS1 |
CNTFR antisense RNA 1 |
8800 |
0.1 |
| chr2_231692712_231693467 | 0.21 |
ITM2C |
integral membrane protein 2C |
36265 |
0.14 |
| chr1_110518577_110518847 | 0.21 |
AHCYL1 |
adenosylhomocysteinase-like 1 |
8596 |
0.18 |
| chr4_25031967_25032301 | 0.21 |
LGI2 |
leucine-rich repeat LGI family, member 2 |
153 |
0.97 |
| chr5_167719768_167719973 | 0.21 |
WWC1 |
WW and C2 domain containing 1 |
715 |
0.76 |
| chr19_39466589_39467071 | 0.21 |
FBXO17 |
F-box protein 17 |
280 |
0.84 |
| chr2_1597607_1597758 | 0.21 |
AC144450.1 |
|
26203 |
0.23 |
| chr6_129203821_129204789 | 0.21 |
LAMA2 |
laminin, alpha 2 |
37 |
0.99 |
| chr4_175135564_175135744 | 0.21 |
FBXO8 |
F-box protein 8 |
69160 |
0.12 |
| chr17_47091251_47091425 | 0.21 |
RP11-501C14.6 |
|
251 |
0.85 |
| chr2_103104653_103104941 | 0.21 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
15035 |
0.18 |
| chr14_71022448_71023169 | 0.21 |
ADAM20 |
ADAM metallopeptidase domain 20 |
21076 |
0.17 |
| chr9_116356336_116356765 | 0.21 |
RGS3 |
regulator of G-protein signaling 3 |
784 |
0.67 |
| chr19_1241899_1242560 | 0.21 |
ATP5D |
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
410 |
0.68 |
| chr7_22538999_22539909 | 0.21 |
STEAP1B |
STEAP family member 1B |
342 |
0.91 |
| chr3_45076964_45077591 | 0.21 |
ENSG00000252410 |
. |
1197 |
0.45 |
| chr4_81189244_81189728 | 0.21 |
FGF5 |
fibroblast growth factor 5 |
1693 |
0.45 |
| chr3_55516545_55517089 | 0.21 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1396 |
0.5 |
| chr1_50061585_50061736 | 0.21 |
AGBL4-IT1 |
AGBL4 intronic transcript 1 (non-protein coding) |
123903 |
0.06 |
| chr22_47149775_47150260 | 0.21 |
CTA-29F11.1 |
|
8443 |
0.18 |
| chr17_46674549_46675551 | 0.21 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
1249 |
0.2 |
| chr20_43970794_43971114 | 0.21 |
SDC4 |
syndecan 4 |
6110 |
0.13 |
| chr19_12949779_12950060 | 0.21 |
MAST1 |
microtubule associated serine/threonine kinase 1 |
571 |
0.48 |
| chr3_184209968_184210530 | 0.21 |
EIF2B5-IT1 |
EIF2B5 intronic transcript 1 (non-protein coding) |
12652 |
0.18 |
| chr11_22214479_22215097 | 0.20 |
ANO5 |
anoctamin 5 |
66 |
0.98 |
| chr1_33546740_33547295 | 0.20 |
ADC |
arginine decarboxylase |
249 |
0.93 |
| chr9_139025166_139025528 | 0.20 |
C9orf69 |
chromosome 9 open reading frame 69 |
14616 |
0.18 |
| chr11_10757586_10757776 | 0.20 |
ENSG00000206858 |
. |
10231 |
0.15 |
| chr17_881575_881780 | 0.20 |
NXN |
nucleoredoxin |
1333 |
0.34 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.5 | GO:0060068 | vagina development(GO:0060068) |
| 0.1 | 0.5 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.2 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.1 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.2 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0060761 | negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.4 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0048243 | regulation of norepinephrine secretion(GO:0014061) norepinephrine secretion(GO:0048243) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.2 | GO:0038061 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) mesenchymal to epithelial transition(GO:0060231) renal vesicle morphogenesis(GO:0072077) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.0 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0022839 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |