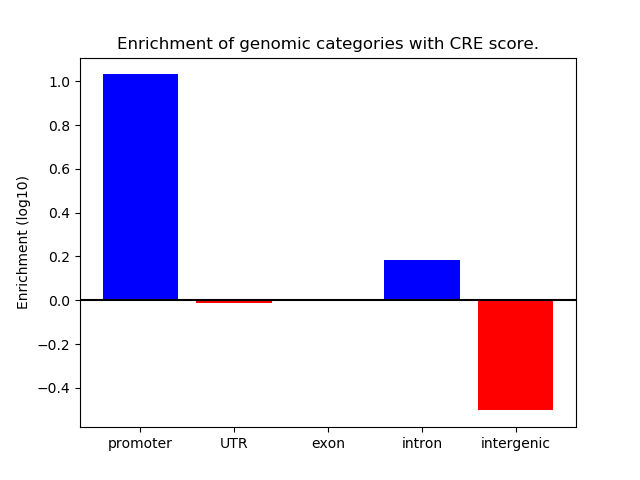
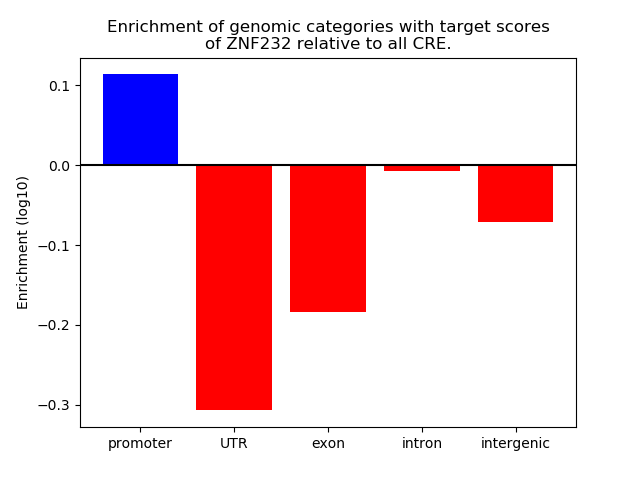
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
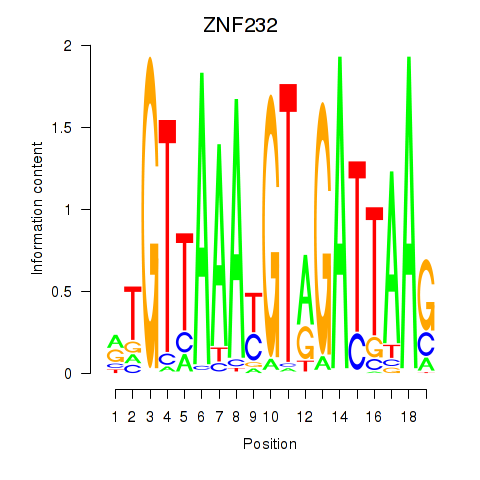
Results for ZNF232
Z-value: 0.35
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
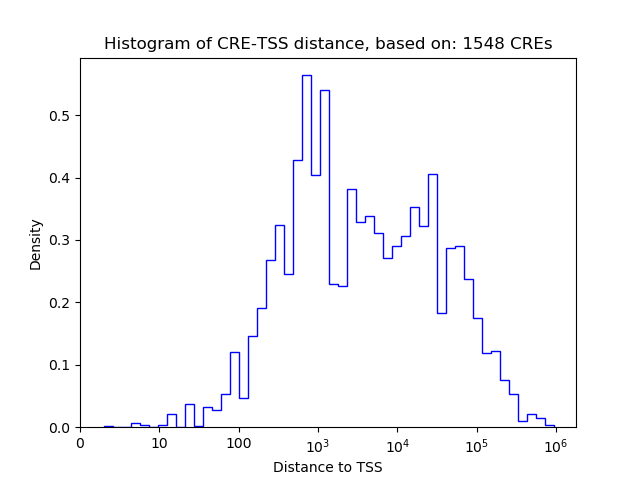
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_5025984_5026458 | ZNF232 | 176 | 0.905187 | -0.36 | 3.4e-01 | Click! |
| chr17_5025740_5025978 | ZNF232 | 538 | 0.633731 | -0.30 | 4.4e-01 | Click! |
| chr17_5026549_5026700 | ZNF232 | 227 | 0.873778 | -0.28 | 4.6e-01 | Click! |
| chr17_5014924_5015118 | ZNF232 | 115 | 0.617372 | 0.17 | 6.6e-01 | Click! |
Activity of the ZNF232 motif across conditions
Conditions sorted by the z-value of the ZNF232 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
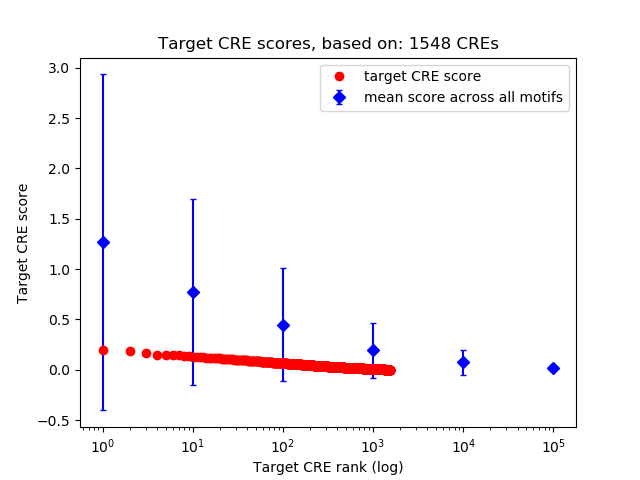
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_130883505_130884015 | 0.20 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
15034 |
0.29 |
| chr12_94544055_94544362 | 0.19 |
PLXNC1 |
plexin C1 |
1709 |
0.42 |
| chr2_113432534_113432708 | 0.17 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
27342 |
0.16 |
| chr1_111416320_111416529 | 0.15 |
CD53 |
CD53 molecule |
648 |
0.73 |
| chr6_105405546_105405738 | 0.15 |
LIN28B |
lin-28 homolog B (C. elegans) |
719 |
0.75 |
| chr10_11206019_11206557 | 0.15 |
CELF2 |
CUGBP, Elav-like family member 2 |
705 |
0.72 |
| chr3_151961203_151961354 | 0.14 |
MBNL1 |
muscleblind-like splicing regulator 1 |
24551 |
0.2 |
| chr2_160711557_160711708 | 0.14 |
LY75 |
lymphocyte antigen 75 |
49561 |
0.13 |
| chr20_52538846_52539125 | 0.14 |
AC005220.3 |
|
17714 |
0.25 |
| chr7_150148744_150149359 | 0.13 |
GIMAP8 |
GTPase, IMAP family member 8 |
1333 |
0.41 |
| chr3_46995163_46995374 | 0.13 |
CCDC12 |
coiled-coil domain containing 12 |
23002 |
0.14 |
| chr3_19189181_19189385 | 0.13 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
663 |
0.83 |
| chr19_35927435_35927735 | 0.12 |
FFAR2 |
free fatty acid receptor 2 |
11618 |
0.11 |
| chr3_108205436_108205587 | 0.12 |
MYH15 |
myosin, heavy chain 15 |
42658 |
0.16 |
| chr9_102972411_102972625 | 0.12 |
ENSG00000239908 |
. |
107677 |
0.06 |
| chr15_34609178_34609329 | 0.12 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
1727 |
0.23 |
| chr12_80265785_80265936 | 0.12 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
792 |
0.63 |
| chr18_18685949_18686100 | 0.12 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
5788 |
0.23 |
| chr1_40844474_40844625 | 0.11 |
SMAP2 |
small ArfGAP2 |
4229 |
0.21 |
| chr2_43232738_43232956 | 0.11 |
ENSG00000207087 |
. |
85785 |
0.1 |
| chr10_14603489_14603761 | 0.11 |
FAM107B |
family with sequence similarity 107, member B |
5446 |
0.27 |
| chr7_50350563_50350714 | 0.11 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
2320 |
0.42 |
| chr3_30715119_30715270 | 0.11 |
RP11-1024P17.1 |
|
23959 |
0.25 |
| chr1_50575996_50576351 | 0.11 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
795 |
0.71 |
| chr22_30234443_30234651 | 0.11 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
282 |
0.9 |
| chr3_19188036_19188187 | 0.11 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
1835 |
0.51 |
| chr14_92340111_92340262 | 0.11 |
FBLN5 |
fibulin 5 |
4200 |
0.23 |
| chr8_92051530_92051722 | 0.10 |
TMEM55A |
transmembrane protein 55A |
1437 |
0.31 |
| chr7_15620685_15620964 | 0.10 |
AGMO |
alkylglycerol monooxygenase |
19184 |
0.26 |
| chr13_27825183_27825382 | 0.10 |
RPL21 |
ribosomal protein L21 |
164 |
0.92 |
| chr10_134150291_134150926 | 0.10 |
LRRC27 |
leucine rich repeat containing 27 |
236 |
0.93 |
| chr12_116716284_116716544 | 0.10 |
MED13L |
mediator complex subunit 13-like |
1271 |
0.52 |
| chr1_24828022_24828241 | 0.10 |
RCAN3 |
RCAN family member 3 |
1256 |
0.44 |
| chr2_66673742_66673893 | 0.10 |
MEIS1 |
Meis homeobox 1 |
3720 |
0.19 |
| chr5_126094487_126094638 | 0.09 |
ENSG00000252185 |
. |
3453 |
0.28 |
| chr1_40848271_40848430 | 0.09 |
SMAP2 |
small ArfGAP2 |
8030 |
0.18 |
| chr13_41553714_41553865 | 0.09 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
2629 |
0.29 |
| chr22_30268883_30269034 | 0.09 |
MTMR3 |
myotubularin related protein 3 |
10186 |
0.16 |
| chrX_130875960_130876221 | 0.09 |
ENSG00000200587 |
. |
196894 |
0.03 |
| chr5_39199802_39200193 | 0.09 |
FYB |
FYN binding protein |
3132 |
0.36 |
| chr18_9102187_9102500 | 0.09 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
285 |
0.9 |
| chr6_149866537_149867090 | 0.09 |
PPIL4 |
peptidylprolyl isomerase (cyclophilin)-like 4 |
361 |
0.81 |
| chr2_232650134_232650432 | 0.09 |
PDE6D |
phosphodiesterase 6D, cGMP-specific, rod, delta |
699 |
0.46 |
| chrX_123096191_123096863 | 0.09 |
STAG2 |
stromal antigen 2 |
493 |
0.85 |
| chr1_78443552_78443956 | 0.09 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
991 |
0.46 |
| chr1_205283585_205283736 | 0.09 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
7223 |
0.17 |
| chr6_110796871_110797126 | 0.09 |
SLC22A16 |
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
601 |
0.69 |
| chr17_56412846_56413076 | 0.09 |
ENSG00000264399 |
. |
422 |
0.69 |
| chr14_91709116_91709392 | 0.09 |
CTD-2547L24.3 |
HCG1816139; Uncharacterized protein |
25 |
0.97 |
| chr8_53627211_53627825 | 0.09 |
RB1CC1 |
RB1-inducible coiled-coil 1 |
492 |
0.84 |
| chrX_77166269_77166668 | 0.09 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
269 |
0.89 |
| chr6_26198173_26198564 | 0.09 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr5_98362367_98362646 | 0.09 |
ENSG00000200351 |
. |
90055 |
0.09 |
| chr22_38784304_38784455 | 0.08 |
RP1-5O6.4 |
|
483 |
0.72 |
| chr5_49735948_49736099 | 0.08 |
EMB |
embigin |
1161 |
0.67 |
| chr10_74146912_74147063 | 0.08 |
ENSG00000202513 |
. |
4735 |
0.23 |
| chr1_158773010_158773161 | 0.08 |
OR6N2 |
olfactory receptor, family 6, subfamily N, member 2 |
25660 |
0.13 |
| chr1_169675171_169675322 | 0.08 |
SELL |
selectin L |
5593 |
0.21 |
| chr12_68383559_68383710 | 0.08 |
IFNG-AS1 |
IFNG antisense RNA 1 |
325 |
0.94 |
| chr17_30471048_30471199 | 0.08 |
AC090616.2 |
Uncharacterized protein |
969 |
0.38 |
| chr1_231665268_231665468 | 0.08 |
TSNAX |
translin-associated factor X |
967 |
0.41 |
| chr12_6446702_6447173 | 0.08 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
4077 |
0.14 |
| chr7_151039214_151039888 | 0.08 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
653 |
0.66 |
| chrX_78417451_78417673 | 0.08 |
GPR174 |
G protein-coupled receptor 174 |
8907 |
0.32 |
| chrX_77154989_77155623 | 0.08 |
COX7B |
cytochrome c oxidase subunit VIIb |
371 |
0.82 |
| chr1_22401714_22401865 | 0.08 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
16099 |
0.13 |
| chrX_8150685_8150836 | 0.08 |
VCX2 |
variable charge, X-linked 2 |
11452 |
0.28 |
| chr2_10472002_10472153 | 0.08 |
HPCAL1 |
hippocalcin-like 1 |
28251 |
0.15 |
| chr8_124049779_124050367 | 0.07 |
TBC1D31 |
TBC1 domain family, member 31 |
4135 |
0.17 |
| chr14_90084669_90085459 | 0.07 |
FOXN3 |
forkhead box N3 |
410 |
0.78 |
| chr12_51321341_51321492 | 0.07 |
METTL7A |
methyltransferase like 7A |
2882 |
0.21 |
| chr19_55020488_55020639 | 0.07 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
6440 |
0.11 |
| chr1_38314226_38314377 | 0.07 |
MTF1 |
metal-regulatory transcription factor 1 |
10991 |
0.09 |
| chr9_67232492_67232789 | 0.07 |
ENSG00000265601 |
. |
7188 |
0.28 |
| chr15_45878869_45879476 | 0.07 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
149 |
0.75 |
| chr6_109416364_109416609 | 0.07 |
CEP57L1 |
centrosomal protein 57kDa-like 1 |
95 |
0.86 |
| chr3_148995184_148995347 | 0.07 |
RP11-206M11.7 |
|
7304 |
0.21 |
| chr7_30067058_30067509 | 0.07 |
PLEKHA8 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
695 |
0.49 |
| chr9_88420732_88421441 | 0.07 |
AGTPBP1 |
ATP/GTP binding protein 1 |
64142 |
0.15 |
| chr3_18478950_18479101 | 0.07 |
SATB1 |
SATB homeobox 1 |
1159 |
0.53 |
| chr13_21649079_21649364 | 0.07 |
LATS2 |
large tumor suppressor kinase 2 |
13535 |
0.15 |
| chr1_169679248_169679951 | 0.07 |
SELL |
selectin L |
1240 |
0.48 |
| chr11_35066286_35066437 | 0.07 |
PDHX |
pyruvate dehydrogenase complex, component X |
67030 |
0.1 |
| chr9_273424_273720 | 0.07 |
DOCK8 |
dedicator of cytokinesis 8 |
502 |
0.5 |
| chrX_10066246_10067411 | 0.07 |
WWC3 |
WWC family member 3 |
35329 |
0.15 |
| chr4_109901499_109901650 | 0.07 |
COL25A1 |
collagen, type XXV, alpha 1 |
156199 |
0.04 |
| chr5_1345378_1345625 | 0.07 |
CLPTM1L |
CLPTM1-like |
287 |
0.91 |
| chr13_50715497_50715648 | 0.07 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
59265 |
0.12 |
| chr11_121340288_121340439 | 0.07 |
RP11-730K11.1 |
|
16641 |
0.23 |
| chr19_39341392_39342222 | 0.07 |
HNRNPL |
heterogeneous nuclear ribonucleoprotein L |
12 |
0.94 |
| chr1_234614976_234615210 | 0.07 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
244 |
0.94 |
| chr20_57738915_57739179 | 0.07 |
ZNF831 |
zinc finger protein 831 |
27028 |
0.21 |
| chr10_120489799_120489950 | 0.07 |
CACUL1 |
CDK2-associated, cullin domain 1 |
24884 |
0.21 |
| chr14_22321778_22321996 | 0.07 |
ENSG00000222776 |
. |
73102 |
0.11 |
| chr20_33733027_33733178 | 0.07 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
143 |
0.94 |
| chr3_21788653_21788804 | 0.07 |
ZNF385D |
zinc finger protein 385D |
4199 |
0.35 |
| chr2_66806799_66806950 | 0.07 |
MEIS1 |
Meis homeobox 1 |
70815 |
0.12 |
| chr21_32556834_32556985 | 0.07 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
54370 |
0.16 |
| chr7_76236072_76236223 | 0.07 |
POMZP3 |
POM121 and ZP3 fusion |
11470 |
0.2 |
| chr11_33907227_33907391 | 0.07 |
AC132216.1 |
HCG1785179; PRO1787; Uncharacterized protein |
5120 |
0.22 |
| chr4_94755757_94755908 | 0.07 |
ATOH1 |
atonal homolog 1 (Drosophila) |
5790 |
0.27 |
| chr1_247095902_247096053 | 0.07 |
AHCTF1 |
AT hook containing transcription factor 1 |
697 |
0.73 |
| chr12_32115176_32115327 | 0.07 |
KIAA1551 |
KIAA1551 |
172 |
0.96 |
| chr12_29435079_29435230 | 0.07 |
RP11-996F15.2 |
|
35627 |
0.16 |
| chr16_12523462_12523613 | 0.06 |
RP11-165M1.2 |
|
56857 |
0.11 |
| chr12_11962518_11962720 | 0.06 |
ETV6 |
ets variant 6 |
57184 |
0.16 |
| chr1_202937217_202937368 | 0.06 |
CYB5R1 |
cytochrome b5 reductase 1 |
884 |
0.46 |
| chr10_65024685_65024930 | 0.06 |
JMJD1C |
jumonji domain containing 1C |
4019 |
0.32 |
| chr19_54871270_54871421 | 0.06 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
1211 |
0.31 |
| chr20_62535769_62535920 | 0.06 |
ENSG00000199805 |
. |
4464 |
0.08 |
| chr5_143267127_143267278 | 0.06 |
HMHB1 |
histocompatibility (minor) HB-1 |
75476 |
0.12 |
| chr4_73232259_73232410 | 0.06 |
RP11-373J21.1 |
|
43589 |
0.21 |
| chr1_198620819_198621442 | 0.06 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
12838 |
0.23 |
| chr20_48321626_48321777 | 0.06 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
8714 |
0.2 |
| chr9_80850286_80850511 | 0.06 |
CEP78 |
centrosomal protein 78kDa |
580 |
0.83 |
| chr11_65408305_65408462 | 0.06 |
SIPA1 |
signal-induced proliferation-associated 1 |
84 |
0.92 |
| chr16_74331532_74331708 | 0.06 |
PSMD7 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
893 |
0.6 |
| chr9_67087582_67087733 | 0.06 |
ENSG00000265285 |
. |
7276 |
0.31 |
| chr5_87798376_87798680 | 0.06 |
TMEM161B-AS1 |
TMEM161B antisense RNA 1 |
76016 |
0.11 |
| chr1_72347063_72347214 | 0.06 |
NEGR1-IT1 |
NEGR1 intronic transcript 1 (non-protein coding) |
44443 |
0.21 |
| chr1_154378708_154379562 | 0.06 |
RP11-350G8.5 |
|
95 |
0.93 |
| chrX_131158192_131158580 | 0.06 |
MST4 |
Serine/threonine-protein kinase MST4 |
750 |
0.71 |
| chr1_160548800_160549253 | 0.06 |
CD84 |
CD84 molecule |
237 |
0.91 |
| chr14_100873124_100873275 | 0.06 |
RP11-362L22.1 |
|
43 |
0.96 |
| chr7_30323590_30323942 | 0.06 |
ZNRF2 |
zinc and ring finger 2 |
157 |
0.96 |
| chr17_8867884_8868296 | 0.06 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
934 |
0.65 |
| chr15_72665455_72665606 | 0.06 |
HEXA |
hexosaminidase A (alpha polypeptide) |
2655 |
0.21 |
| chr8_8861066_8861441 | 0.06 |
ERI1 |
exoribonuclease 1 |
899 |
0.57 |
| chr2_101921816_101922067 | 0.06 |
RNF149 |
ring finger protein 149 |
3217 |
0.21 |
| chr2_170591300_170591489 | 0.06 |
KLHL23 |
kelch-like family member 23 |
668 |
0.64 |
| chr19_9545439_9546179 | 0.06 |
ZNF266 |
zinc finger protein 266 |
60 |
0.97 |
| chr15_69707071_69708026 | 0.06 |
RP11-253M7.1 |
|
180 |
0.86 |
| chr18_2659017_2659168 | 0.06 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
3206 |
0.21 |
| chr10_134068601_134068752 | 0.06 |
STK32C |
serine/threonine kinase 32C |
52042 |
0.12 |
| chr13_45058136_45058287 | 0.06 |
TSC22D1 |
TSC22 domain family, member 1 |
9825 |
0.29 |
| chr2_190133275_190133426 | 0.06 |
ENSG00000266817 |
. |
43137 |
0.17 |
| chr15_99395675_99396414 | 0.06 |
IGF1R |
insulin-like growth factor 1 receptor |
37526 |
0.16 |
| chr4_141075455_141075656 | 0.06 |
MAML3 |
mastermind-like 3 (Drosophila) |
217 |
0.95 |
| chr1_42230218_42230369 | 0.06 |
ENSG00000264896 |
. |
5481 |
0.3 |
| chr3_127842347_127842602 | 0.06 |
RUVBL1 |
RuvB-like AAA ATPase 1 |
169 |
0.94 |
| chr1_232726179_232726330 | 0.06 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
28950 |
0.24 |
| chr4_38857928_38858431 | 0.06 |
TLR6 |
toll-like receptor 6 |
258 |
0.48 |
| chr14_45719805_45719966 | 0.06 |
MIS18BP1 |
MIS18 binding protein 1 |
2495 |
0.3 |
| chr20_31484373_31484524 | 0.06 |
EFCAB8 |
EF-hand calcium binding domain 8 |
15011 |
0.18 |
| chr2_54221778_54222027 | 0.06 |
ACYP2 |
acylphosphatase 2, muscle type |
23657 |
0.18 |
| chr8_59977467_59977618 | 0.05 |
RP11-25K19.1 |
|
54057 |
0.13 |
| chr6_106534568_106534850 | 0.05 |
PRDM1 |
PR domain containing 1, with ZNF domain |
514 |
0.83 |
| chr3_14557562_14557713 | 0.05 |
ENSG00000207163 |
. |
14958 |
0.22 |
| chr9_31422111_31422262 | 0.05 |
ENSG00000211510 |
. |
647960 |
0.0 |
| chr17_60140816_60141064 | 0.05 |
MED13 |
mediator complex subunit 13 |
1703 |
0.35 |
| chr4_48342236_48343218 | 0.05 |
SLAIN2 |
SLAIN motif family, member 2 |
612 |
0.8 |
| chrX_24043946_24044693 | 0.05 |
KLHL15 |
kelch-like family member 15 |
984 |
0.59 |
| chrY_2482600_2482989 | 0.05 |
ENSG00000251841 |
. |
169996 |
0.04 |
| chr9_42914569_42914720 | 0.05 |
AC129778.2 |
|
55559 |
0.15 |
| chr6_111202605_111202756 | 0.05 |
AMD1 |
adenosylmethionine decarboxylase 1 |
5902 |
0.17 |
| chr7_114565895_114566046 | 0.05 |
MDFIC |
MyoD family inhibitor domain containing |
2839 |
0.42 |
| chr1_159925204_159925355 | 0.05 |
SLAMF9 |
SLAM family member 9 |
1235 |
0.32 |
| chr8_117664337_117664488 | 0.05 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
103611 |
0.07 |
| chr3_119811066_119811419 | 0.05 |
GSK3B |
glycogen synthase kinase 3 beta |
1271 |
0.47 |
| chr10_65028344_65028625 | 0.05 |
JMJD1C |
jumonji domain containing 1C |
342 |
0.93 |
| chr10_111769664_111769909 | 0.05 |
ADD3 |
adducin 3 (gamma) |
2064 |
0.33 |
| chr1_167298272_167298450 | 0.05 |
POU2F1 |
POU class 2 homeobox 1 |
80 |
0.98 |
| chr20_5588883_5589774 | 0.05 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
2344 |
0.4 |
| chr2_24577017_24577377 | 0.05 |
ITSN2 |
intersectin 2 |
5981 |
0.25 |
| chr6_57039244_57039395 | 0.05 |
RP11-203B9.4 |
|
285 |
0.85 |
| chr7_130789448_130789720 | 0.05 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
2205 |
0.33 |
| chr10_124713000_124713296 | 0.05 |
C10orf88 |
chromosome 10 open reading frame 88 |
771 |
0.61 |
| chr2_232526451_232526617 | 0.05 |
ENSG00000239202 |
. |
15550 |
0.16 |
| chrX_78619893_78620072 | 0.05 |
ITM2A |
integral membrane protein 2A |
2874 |
0.42 |
| chr3_20145563_20145846 | 0.05 |
ENSG00000266745 |
. |
33353 |
0.15 |
| chr15_50568045_50568196 | 0.05 |
HDC |
histidine decarboxylase |
9897 |
0.15 |
| chr16_87096172_87096484 | 0.05 |
RP11-899L11.3 |
|
153193 |
0.04 |
| chr8_52783217_52783484 | 0.05 |
PCMTD1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
25045 |
0.15 |
| chr13_110810904_110811055 | 0.05 |
ENSG00000265885 |
. |
60481 |
0.14 |
| chr5_69586650_69586865 | 0.05 |
GTF2H2B |
general transcription factor IIH, polypeptide 2B (pseudogene) |
124422 |
0.06 |
| chr5_15718526_15718677 | 0.05 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
102510 |
0.08 |
| chr16_21612011_21612297 | 0.05 |
METTL9 |
methyltransferase like 9 |
1275 |
0.35 |
| chr11_111444678_111444829 | 0.05 |
SIK2 |
salt-inducible kinase 2 |
28362 |
0.11 |
| chr5_69882192_69882402 | 0.05 |
RP11-497H16.7 |
|
113022 |
0.07 |
| chr11_18127000_18127190 | 0.05 |
SAAL1 |
serum amyloid A-like 1 |
471 |
0.74 |
| chr1_101650998_101651149 | 0.05 |
ENSG00000238296 |
. |
43248 |
0.11 |
| chrX_55479729_55479880 | 0.05 |
MAGEH1 |
melanoma antigen family H, 1 |
1266 |
0.4 |
| chr5_69006919_69007136 | 0.05 |
GUSBP3 |
glucuronidase, beta pseudogene 3 |
686 |
0.78 |
| chr6_41042589_41042740 | 0.05 |
NFYA |
nuclear transcription factor Y, alpha |
1942 |
0.21 |
| chr4_143224062_143224213 | 0.05 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
2848 |
0.43 |
| chr10_27529985_27530175 | 0.05 |
ACBD5 |
acyl-CoA binding domain containing 5 |
301 |
0.85 |
| chr14_78081630_78082591 | 0.05 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
1006 |
0.56 |
| chr5_70014898_70015115 | 0.05 |
SERF1A |
small EDRK-rich factor 1A (telomeric) |
181486 |
0.03 |
| chr11_102320072_102320223 | 0.05 |
TMEM123 |
transmembrane protein 123 |
36 |
0.97 |
| chr5_95144147_95144466 | 0.05 |
GLRX |
glutaredoxin (thioltransferase) |
14109 |
0.14 |
| chr5_180671226_180671377 | 0.05 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
92 |
0.87 |
| chr13_74067154_74067305 | 0.05 |
ENSG00000206697 |
. |
265945 |
0.02 |
| chr2_159074611_159074762 | 0.05 |
CCDC148-AS1 |
CCDC148 antisense RNA 1 |
51524 |
0.15 |
| chr12_120974205_120974387 | 0.05 |
RNF10 |
ring finger protein 10 |
1067 |
0.34 |
| chr2_265229_265468 | 0.05 |
ACP1 |
acid phosphatase 1, soluble |
422 |
0.54 |
| chr2_45004927_45005078 | 0.05 |
ENSG00000252896 |
. |
4267 |
0.35 |
| chr13_43566462_43567025 | 0.05 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
358 |
0.92 |
| chr21_16579081_16579232 | 0.05 |
NRIP1 |
nuclear receptor interacting protein 1 |
141835 |
0.05 |
| chr5_70586170_70586382 | 0.05 |
PMCHL2 |
pro-melanin-concentrating hormone-like 2, pseudogene |
85381 |
0.09 |
| chr2_114655602_114655753 | 0.05 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
7502 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |