Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
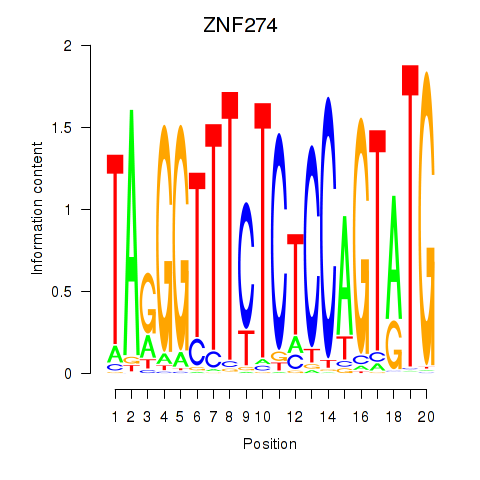
Results for ZNF274
Z-value: 8.50
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_58689540_58689691 | ZNF274 | 4781 | 0.148182 | 0.86 | 2.6e-03 | Click! |
| chr19_58709074_58709237 | ZNF274 | 14292 | 0.118807 | 0.84 | 5.0e-03 | Click! |
| chr19_58703717_58703868 | ZNF274 | 8929 | 0.128998 | 0.80 | 9.4e-03 | Click! |
| chr19_58708556_58708707 | ZNF274 | 13768 | 0.119717 | 0.74 | 2.4e-02 | Click! |
| chr19_58705561_58705712 | ZNF274 | 10773 | 0.125116 | 0.72 | 2.9e-02 | Click! |
Activity of the ZNF274 motif across conditions
Conditions sorted by the z-value of the ZNF274 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
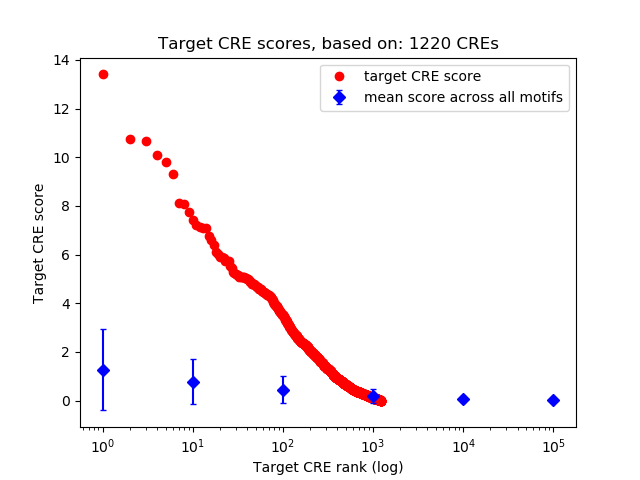
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_99668638_99669334 | 13.40 |
ZNF3 |
zinc finger protein 3 |
6224 |
0.07 |
| chr12_133779073_133779476 | 10.76 |
AC226150.4 |
Uncharacterized protein |
8498 |
0.13 |
| chr19_12061015_12061385 | 10.67 |
ZNF700 |
zinc finger protein 700 |
3631 |
0.13 |
| chr19_37619154_37619493 | 10.09 |
ZNF420 |
zinc finger protein 420 |
254 |
0.89 |
| chr19_37903473_37903682 | 9.82 |
ZNF569 |
zinc finger protein 569 |
32284 |
0.12 |
| chr19_12637235_12637523 | 9.30 |
CTD-2192J16.21 |
|
6408 |
0.12 |
| chr19_12637638_12637867 | 8.14 |
CTD-2192J16.21 |
|
6035 |
0.13 |
| chr19_44418001_44418215 | 8.10 |
ZNF45 |
zinc finger protein 45 |
5783 |
0.13 |
| chr7_99128907_99129569 | 7.74 |
FAM200A |
family with sequence similarity 200, member A |
20519 |
0.09 |
| chr19_52569993_52570228 | 7.42 |
ZNF841 |
zinc finger protein 841 |
676 |
0.53 |
| chr19_44352263_44352470 | 7.21 |
ZNF283 |
zinc finger protein 283 |
20012 |
0.11 |
| chr16_71482989_71483214 | 7.13 |
ZNF23 |
zinc finger protein 23 |
4775 |
0.14 |
| chr19_37238828_37239050 | 7.11 |
ZNF850 |
zinc finger protein 850 |
24777 |
0.12 |
| chr19_37619538_37619802 | 7.09 |
ZNF420 |
zinc finger protein 420 |
601 |
0.66 |
| chr8_146156057_146156445 | 6.76 |
ZNF16 |
zinc finger protein 16 |
19730 |
0.14 |
| chr16_3339867_3340112 | 6.61 |
ZNF263 |
zinc finger protein 263 |
223 |
0.87 |
| chr19_57066223_57066374 | 6.37 |
AC007228.11 |
|
12482 |
0.12 |
| chr8_146156566_146156813 | 6.09 |
ZNF16 |
zinc finger protein 16 |
19292 |
0.14 |
| chr12_133780561_133780891 | 6.01 |
AC226150.4 |
Uncharacterized protein |
7046 |
0.13 |
| chr8_146067350_146067618 | 5.91 |
COMMD5 |
COMM domain containing 5 |
10558 |
0.11 |
| chr19_37382488_37382676 | 5.89 |
ZNF345 |
zinc finger protein 345 |
18982 |
0.12 |
| chr19_52569166_52569317 | 5.87 |
ZNF432 |
zinc finger protein 432 |
1460 |
0.22 |
| chr6_27419133_27419361 | 5.74 |
ZNF184 |
zinc finger protein 184 |
21213 |
0.2 |
| chr19_53344303_53344454 | 5.73 |
ZNF468 |
zinc finger protein 468 |
8505 |
0.16 |
| chr19_57089373_57089524 | 5.72 |
ZNF470 |
zinc finger protein 470 |
3432 |
0.16 |
| chr19_38229308_38229579 | 5.54 |
ZNF607 |
zinc finger protein 607 |
18752 |
0.15 |
| chr4_435829_435993 | 5.47 |
ENSG00000252150 |
. |
25514 |
0.13 |
| chr19_12383361_12383540 | 5.28 |
ZNF44 |
zinc finger protein 44 |
21832 |
0.15 |
| chr19_12187967_12188204 | 5.26 |
ENSG00000252546 |
. |
7106 |
0.09 |
| chr19_11980011_11980162 | 5.21 |
ZNF439 |
zinc finger protein 439 |
3314 |
0.16 |
| chr19_58058409_58058690 | 5.18 |
ZNF550 |
zinc finger protein 550 |
812 |
0.44 |
| chr19_58773764_58774018 | 5.16 |
CTD-3138B18.6 |
|
4110 |
0.12 |
| chr19_37904473_37904624 | 5.10 |
ZNF569 |
zinc finger protein 569 |
31313 |
0.12 |
| chr19_58945338_58945731 | 5.09 |
ZNF132 |
zinc finger protein 132 |
6055 |
0.08 |
| chr19_7083204_7083430 | 5.08 |
ZNF557 |
zinc finger protein 557 |
13601 |
0.13 |
| chr19_52888445_52888596 | 5.08 |
ZNF528 |
zinc finger protein 528 |
12582 |
0.11 |
| chr19_58437657_58437808 | 5.06 |
ZNF418 |
zinc finger protein 418 |
7599 |
0.11 |
| chr19_44661345_44661496 | 5.06 |
ZNF226 |
zinc finger protein 226 |
7806 |
0.12 |
| chr12_133779730_133779883 | 5.04 |
AC226150.4 |
Uncharacterized protein |
7966 |
0.13 |
| chr7_6731088_6731239 | 5.01 |
ENSG00000265245 |
. |
445 |
0.74 |
| chr19_57088795_57088946 | 4.98 |
ZNF470 |
zinc finger protein 470 |
2854 |
0.18 |
| chr19_58265433_58265584 | 4.94 |
ZNF776 |
zinc finger protein 776 |
3336 |
0.12 |
| chr19_58084288_58084439 | 4.88 |
ZNF416 |
zinc finger protein 416 |
5932 |
0.1 |
| chr19_53209321_53209472 | 4.88 |
ZNF83 |
zinc finger protein 83 |
15647 |
0.14 |
| chr19_37880364_37880570 | 4.79 |
ZNF527 |
zinc finger protein 527 |
18371 |
0.15 |
| chr19_37367869_37368020 | 4.79 |
ZNF345 |
zinc finger protein 345 |
4344 |
0.16 |
| chr19_58198938_58199089 | 4.78 |
AC004017.1 |
Uncharacterized protein |
5115 |
0.09 |
| chr6_27419825_27419976 | 4.78 |
ZNF184 |
zinc finger protein 184 |
20560 |
0.21 |
| chr19_21365931_21366082 | 4.76 |
ZNF431 |
zinc finger protein 431 |
41126 |
0.16 |
| chr1_247201223_247201374 | 4.71 |
ZNF695 |
zinc finger protein 695 |
29903 |
0.15 |
| chr10_38120780_38120931 | 4.69 |
ENSG00000266800 |
. |
38 |
0.98 |
| chr16_31927650_31927801 | 4.67 |
ZNF267 |
zinc finger protein 267 |
42338 |
0.14 |
| chr19_9406532_9406683 | 4.65 |
CTC-325H20.4 |
|
4416 |
0.13 |
| chr19_12016356_12016507 | 4.59 |
ZNF69 |
zinc finger protein 69 |
17761 |
0.1 |
| chr1_23688765_23688916 | 4.58 |
ZNF436 |
zinc finger protein 436 |
6039 |
0.14 |
| chr7_5104801_5105169 | 4.58 |
RBAK |
RB-associated KRAB zinc finger |
19432 |
0.16 |
| chr19_24309794_24309945 | 4.58 |
ZNF254 |
zinc finger protein 254 |
39874 |
0.2 |
| chr7_99091524_99091729 | 4.52 |
ZNF394 |
zinc finger protein 394 |
6261 |
0.1 |
| chr19_12060636_12060787 | 4.49 |
ZNF700 |
zinc finger protein 700 |
3142 |
0.15 |
| chr2_95881225_95881376 | 4.48 |
ZNF2 |
zinc finger protein 2 |
49738 |
0.11 |
| chr19_19822222_19822373 | 4.47 |
ZNF14 |
zinc finger protein 14 |
21609 |
0.12 |
| chr9_115805477_115805628 | 4.46 |
ZFP37 |
ZFP37 zinc finger protein |
13416 |
0.24 |
| chr5_150275044_150275195 | 4.42 |
ZNF300 |
zinc finger protein 300 |
9232 |
0.19 |
| chr3_44762895_44763046 | 4.41 |
ZNF501 |
zinc finger protein 501 |
8118 |
0.13 |
| chr19_12637984_12638215 | 4.37 |
CTD-2192J16.21 |
|
5688 |
0.13 |
| chr12_133634115_133634266 | 4.36 |
ZNF84 |
zinc finger protein 84 |
16319 |
0.11 |
| chr19_40581075_40581226 | 4.35 |
ZNF780A |
zinc finger protein 780A |
14992 |
0.13 |
| chr19_12256353_12256531 | 4.35 |
ZNF625 |
zinc finger protein 625 |
2175 |
0.18 |
| chr5_178506733_178506884 | 4.33 |
ZNF354C |
zinc finger protein 354C |
19392 |
0.2 |
| chr19_58489988_58490264 | 4.32 |
C19orf18 |
chromosome 19 open reading frame 18 |
4224 |
0.13 |
| chr19_58774288_58774477 | 4.31 |
CTD-3138B18.6 |
|
3619 |
0.12 |
| chr19_53668202_53668353 | 4.29 |
CTD-2245F17.2 |
|
2596 |
0.18 |
| chr19_58945059_58945210 | 4.26 |
ZNF132 |
zinc finger protein 132 |
6455 |
0.08 |
| chr19_2917142_2917303 | 4.22 |
ZNF57 |
zinc finger protein 57 |
1779 |
0.23 |
| chr19_11833580_11833731 | 4.19 |
CTC-499B15.6 |
|
7943 |
0.13 |
| chr16_89294257_89294408 | 4.16 |
RP11-46C24.6 |
|
768 |
0.5 |
| chr19_58384818_58384969 | 4.11 |
CTD-2583A14.8 |
|
5353 |
0.11 |
| chr19_2933699_2933850 | 4.07 |
AC006277.2 |
|
6969 |
0.12 |
| chr19_52448304_52448455 | 4.02 |
HCCAT3 |
hepatocellular carcinoma associated transcript 3 (non-protein coding) |
4008 |
0.14 |
| chr19_53668494_53668645 | 3.99 |
CTD-2245F17.2 |
|
2888 |
0.17 |
| chr19_12059575_12059873 | 3.95 |
ZNF700 |
zinc finger protein 700 |
2155 |
0.19 |
| chr7_5104239_5104390 | 3.94 |
RBAK |
RB-associated KRAB zinc finger |
18761 |
0.16 |
| chr16_75203576_75203727 | 3.90 |
ZFP1 |
ZFP1 zinc finger protein |
20702 |
0.13 |
| chr19_9868703_9868854 | 3.89 |
ZNF846 |
zinc finger protein 846 |
10203 |
0.12 |
| chr16_71482550_71482701 | 3.87 |
ZNF23 |
zinc finger protein 23 |
5251 |
0.14 |
| chr19_44611379_44611530 | 3.84 |
ZNF225 |
zinc finger protein 225 |
4880 |
0.09 |
| chr19_57037107_57037258 | 3.83 |
ZFP28 |
ZFP28 zinc finger protein |
13135 |
0.12 |
| chr7_6730817_6730968 | 3.81 |
ENSG00000265245 |
. |
716 |
0.56 |
| chr19_44740699_44740850 | 3.75 |
ZNF233 |
zinc finger protein 233 |
13544 |
0.14 |
| chr12_133779970_133780121 | 3.73 |
AC226150.4 |
Uncharacterized protein |
7727 |
0.13 |
| chr19_58579191_58579342 | 3.68 |
ENSG00000243642 |
. |
2072 |
0.2 |
| chr19_20117293_20117444 | 3.67 |
ZNF682 |
zinc finger protein 682 |
703 |
0.66 |
| chr19_11833005_11833156 | 3.65 |
CTC-499B15.6 |
|
8518 |
0.13 |
| chr19_12016806_12016962 | 3.63 |
ZNF69 |
zinc finger protein 69 |
18214 |
0.1 |
| chr19_21910240_21910391 | 3.59 |
ZNF100 |
zinc finger protein 100 |
23199 |
0.17 |
| chr1_247263983_247264134 | 3.59 |
ZNF669 |
zinc finger protein 669 |
3522 |
0.2 |
| chr19_20308367_20308518 | 3.55 |
CTC-260E6.6 |
|
22786 |
0.16 |
| chr19_52448135_52448286 | 3.52 |
HCCAT3 |
hepatocellular carcinoma associated transcript 3 (non-protein coding) |
4177 |
0.14 |
| chr19_12186683_12186905 | 3.51 |
ENSG00000252546 |
. |
5815 |
0.09 |
| chr19_52394180_52394331 | 3.50 |
ZNF577 |
zinc finger protein 577 |
52 |
0.96 |
| chr19_44418412_44418563 | 3.50 |
ZNF45 |
zinc finger protein 45 |
5404 |
0.13 |
| chr19_37368601_37368893 | 3.49 |
ZNF345 |
zinc finger protein 345 |
5147 |
0.15 |
| chr16_71482161_71482312 | 3.45 |
ZNF23 |
zinc finger protein 23 |
5640 |
0.14 |
| chr19_37240168_37240319 | 3.44 |
ZNF850 |
zinc finger protein 850 |
23473 |
0.12 |
| chr19_12384101_12384252 | 3.38 |
ZNF44 |
zinc finger protein 44 |
21452 |
0.15 |
| chr19_40520200_40520351 | 3.33 |
ZNF546 |
zinc finger protein 546 |
17132 |
0.13 |
| chr15_85164723_85164874 | 3.31 |
RP11-182J1.5 |
|
6598 |
0.11 |
| chr19_52537537_52537707 | 3.30 |
ZNF614 |
zinc finger protein 614 |
4129 |
0.13 |
| chr10_38344099_38344250 | 3.28 |
ENSG00000252132 |
. |
11271 |
0.18 |
| chr17_5084544_5084821 | 3.26 |
ZNF594 |
zinc finger protein 594 |
3010 |
0.18 |
| chr19_8922272_8922597 | 3.20 |
CTD-2529P6.3 |
|
9743 |
0.14 |
| chr19_58946276_58946427 | 3.19 |
ZNF132 |
zinc finger protein 132 |
5238 |
0.08 |
| chr19_52570290_52570516 | 3.17 |
ZNF841 |
zinc finger protein 841 |
383 |
0.75 |
| chr19_20003040_20003191 | 3.15 |
ENSG00000265339 |
. |
1147 |
0.37 |
| chr19_44515027_44515178 | 3.14 |
ZNF230 |
zinc finger protein 230 |
4250 |
0.12 |
| chr17_4995332_4995483 | 3.11 |
ZFP3 |
ZFP3 zinc finger protein |
13864 |
0.09 |
| chr19_52887340_52887713 | 3.09 |
ZNF528 |
zinc finger protein 528 |
13576 |
0.11 |
| chr19_52537972_52538123 | 3.03 |
ZNF614 |
zinc finger protein 614 |
4554 |
0.13 |
| chr17_5085116_5085307 | 3.03 |
ZNF594 |
zinc finger protein 594 |
2481 |
0.2 |
| chr19_52537089_52537456 | 3.03 |
ZNF614 |
zinc finger protein 614 |
3779 |
0.14 |
| chr19_58772914_58773185 | 3.02 |
CTD-3138B18.6 |
|
4952 |
0.11 |
| chr16_3432968_3433133 | 2.95 |
MTRNR2L4 |
MT-RNR2-like 4 |
10767 |
0.11 |
| chr19_44514850_44515001 | 2.93 |
ZNF230 |
zinc finger protein 230 |
4073 |
0.12 |
| chr19_58132283_58132434 | 2.91 |
ZNF134 |
zinc finger protein 134 |
6734 |
0.1 |
| chr19_2918008_2918159 | 2.89 |
ZNF57 |
zinc finger protein 57 |
2640 |
0.17 |
| chr19_57889335_57889486 | 2.85 |
ZNF547 |
zinc finger protein 547 |
6413 |
0.08 |
| chr19_11728092_11728444 | 2.84 |
ZNF627 |
zinc finger protein 627 |
19970 |
0.11 |
| chr19_52091081_52091232 | 2.84 |
AC018755.1 |
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
6474 |
0.12 |
| chr19_44418700_44418851 | 2.83 |
ZNF45 |
zinc finger protein 45 |
5116 |
0.13 |
| chr19_53453529_53453680 | 2.81 |
ZNF816 |
zinc finger protein 816 |
7757 |
0.15 |
| chr5_14153831_14154379 | 2.81 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10276 |
0.32 |
| chr19_2853169_2853320 | 2.79 |
AC006130.3 |
|
5470 |
0.11 |
| chr9_99581284_99581435 | 2.78 |
ZNF782 |
zinc finger protein 782 |
35283 |
0.17 |
| chr12_133780930_133781187 | 2.73 |
AC226150.4 |
Uncharacterized protein |
6714 |
0.13 |
| chr7_6731745_6731896 | 2.73 |
ENSG00000265245 |
. |
212 |
0.9 |
| chr19_58944759_58944982 | 2.71 |
ZNF132 |
zinc finger protein 132 |
6719 |
0.08 |
| chr19_40580855_40581006 | 2.70 |
ZNF780A |
zinc finger protein 780A |
15212 |
0.13 |
| chr19_52619612_52619763 | 2.70 |
ENSG00000238630 |
. |
17906 |
0.1 |
| chr19_59073705_59074783 | 2.68 |
AC016629.8 |
|
3652 |
0.09 |
| chr19_44980660_44980811 | 2.67 |
ZNF180 |
zinc finger protein 180 |
23827 |
0.14 |
| chr19_52825941_52826092 | 2.66 |
ZNF610 |
zinc finger protein 610 |
13482 |
0.1 |
| chr19_44351457_44351608 | 2.66 |
ZNF283 |
zinc finger protein 283 |
19178 |
0.11 |
| chr19_57888956_57889107 | 2.65 |
ZNF547 |
zinc finger protein 547 |
6034 |
0.08 |
| chr3_42956766_42956917 | 2.58 |
ZNF662 |
zinc finger protein 662 |
7327 |
0.14 |
| chr19_38126233_38126384 | 2.57 |
ZFP30 |
ZFP30 zinc finger protein |
19322 |
0.14 |
| chr19_52869878_52870029 | 2.56 |
ZNF880 |
zinc finger protein 880 |
3217 |
0.15 |
| chr16_31927384_31927535 | 2.55 |
ZNF267 |
zinc finger protein 267 |
42072 |
0.14 |
| chr19_53958739_53958890 | 2.54 |
ZNF813 |
zinc finger protein 813 |
12175 |
0.13 |
| chr19_12060803_12060954 | 2.53 |
ZNF700 |
zinc finger protein 700 |
3309 |
0.14 |
| chr4_366609_366760 | 2.53 |
ENSG00000207642 |
. |
22738 |
0.14 |
| chr19_57932551_57932702 | 2.50 |
ZNF17 |
zinc finger protein 17 |
7885 |
0.08 |
| chr2_95880533_95880684 | 2.49 |
ZNF2 |
zinc finger protein 2 |
49046 |
0.12 |
| chr10_44052240_44052391 | 2.47 |
ZNF239 |
zinc finger protein 239 |
11592 |
0.2 |
| chr4_367776_367927 | 2.45 |
ENSG00000207642 |
. |
23905 |
0.14 |
| chr19_12574888_12575039 | 2.45 |
CTD-3105H18.7 |
|
13214 |
0.11 |
| chr1_247263735_247263886 | 2.43 |
ZNF669 |
zinc finger protein 669 |
3770 |
0.19 |
| chr19_52496782_52496933 | 2.41 |
ZNF350 |
zinc finger protein 350 |
6748 |
0.12 |
| chr19_11891380_11891531 | 2.40 |
ZNF441 |
zinc finger protein 441 |
13548 |
0.11 |
| chr5_179685390_179685541 | 2.40 |
MAPK9 |
mitogen-activated protein kinase 9 |
22116 |
0.17 |
| chr6_27419414_27419565 | 2.40 |
ZNF184 |
zinc finger protein 184 |
20971 |
0.21 |
| chr5_180276049_180276200 | 2.40 |
ZFP62 |
ZFP62 zinc finger protein |
11539 |
0.16 |
| chr19_44661024_44661182 | 2.39 |
ZNF226 |
zinc finger protein 226 |
8123 |
0.11 |
| chr19_37129764_37129915 | 2.38 |
ZNF461 |
zinc finger protein 461 |
27886 |
0.11 |
| chr17_73126799_73128080 | 2.38 |
NT5C |
5', 3'-nucleotidase, cytosolic |
23 |
0.94 |
| chr19_44514643_44514794 | 2.36 |
ZNF230 |
zinc finger protein 230 |
3866 |
0.12 |
| chr19_53911481_53911632 | 2.36 |
ZNF765 |
zinc finger protein 765 |
13130 |
0.13 |
| chr3_147128128_147129466 | 2.35 |
ZIC1 |
Zic family member 1 |
1626 |
0.36 |
| chr20_31063509_31063841 | 2.35 |
C20orf112 |
chromosome 20 open reading frame 112 |
7599 |
0.19 |
| chr19_57089681_57089933 | 2.35 |
ZNF470 |
zinc finger protein 470 |
3791 |
0.16 |
| chr19_58774047_58774198 | 2.33 |
CTD-3138B18.6 |
|
3879 |
0.12 |
| chr17_5085511_5085662 | 2.33 |
ZNF594 |
zinc finger protein 594 |
2106 |
0.22 |
| chr19_9639881_9640042 | 2.32 |
ZNF426 |
zinc finger protein 426 |
7284 |
0.17 |
| chr19_58579394_58579545 | 2.31 |
ENSG00000243642 |
. |
1869 |
0.22 |
| chr4_437255_437406 | 2.31 |
ENSG00000252150 |
. |
24095 |
0.14 |
| chr2_95814473_95814624 | 2.29 |
ZNF514 |
zinc finger protein 514 |
10534 |
0.17 |
| chr19_9524094_9524245 | 2.29 |
ZNF266 |
zinc finger protein 266 |
5588 |
0.18 |
| chr16_71483294_71483445 | 2.29 |
ZNF23 |
zinc finger protein 23 |
4507 |
0.14 |
| chr19_37037949_37038100 | 2.28 |
ZNF529 |
zinc finger protein 529 |
406 |
0.78 |
| chr19_2933350_2933631 | 2.28 |
AC006277.2 |
|
6685 |
0.12 |
| chr19_52825669_52825820 | 2.27 |
ZNF610 |
zinc finger protein 610 |
13754 |
0.1 |
| chr10_38120611_38120762 | 2.27 |
ENSG00000266800 |
. |
131 |
0.96 |
| chr19_57984721_57984872 | 2.24 |
AC004076.9 |
Uncharacterized protein |
4075 |
0.1 |
| chr5_115790032_115790655 | 2.24 |
CTB-118N6.3 |
|
4917 |
0.26 |
| chr5_178310861_178311012 | 2.22 |
ENSG00000206624 |
. |
780 |
0.59 |
| chr17_15619882_15620033 | 2.21 |
ZNF286A |
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
15444 |
0.12 |
| chr19_58773538_58773689 | 2.20 |
CTD-3138B18.6 |
|
4388 |
0.11 |
| chr19_21607591_21607742 | 2.19 |
ZNF493 |
zinc finger protein 493 |
27698 |
0.19 |
| chr19_2853573_2853724 | 2.19 |
AC006130.3 |
|
5066 |
0.11 |
| chr5_178506431_178506582 | 2.18 |
ZNF354C |
zinc finger protein 354C |
19090 |
0.2 |
| chr1_154912719_154912870 | 2.18 |
PMVK |
phosphomevalonate kinase |
3327 |
0.11 |
| chr5_178310289_178310440 | 2.18 |
ENSG00000206624 |
. |
1352 |
0.38 |
| chr19_19823236_19823387 | 2.12 |
ZNF14 |
zinc finger protein 14 |
20595 |
0.13 |
| chr16_3458666_3458842 | 2.12 |
ZNF174 |
zinc finger protein 174 |
6780 |
0.12 |
| chr19_35449273_35449424 | 2.11 |
ZNF792 |
zinc finger protein 792 |
2259 |
0.22 |
| chr19_52976534_52976685 | 2.10 |
ZNF578 |
zinc finger protein 578 |
19474 |
0.12 |
| chr19_37310288_37310439 | 2.08 |
CTD-2162K18.5 |
|
18252 |
0.12 |
| chr19_11943105_11943256 | 2.08 |
ZNF440 |
zinc finger protein 440 |
2379 |
0.19 |
| chr19_9676750_9676901 | 2.06 |
ZNF121 |
zinc finger protein 121 |
1594 |
0.33 |
| chr12_90484641_90484914 | 2.06 |
ENSG00000252823 |
. |
336941 |
0.01 |
| chr5_178359120_178359271 | 2.05 |
ZNF454 |
zinc finger protein 454 |
8997 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.5 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 1.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.3 | 1.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.3 | 2.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.3 | 0.9 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.2 | 0.5 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.2 | 2.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 1.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 0.9 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 0.2 | GO:0048368 | regulation of mesodermal cell fate specification(GO:0042661) lateral mesoderm development(GO:0048368) regulation of mesoderm development(GO:2000380) |
| 0.2 | 0.5 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.2 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 315.6 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) regulation of nucleic acid-templated transcription(GO:1903506) |
| 0.2 | 0.6 | GO:0042532 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.2 | 1.7 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.2 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.3 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.1 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.2 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0060045 | positive regulation of cardiac muscle tissue growth(GO:0055023) positive regulation of cardiac muscle tissue development(GO:0055025) positive regulation of cardiac muscle cell proliferation(GO:0060045) positive regulation of heart growth(GO:0060421) |
| 0.1 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.4 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.2 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.1 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.7 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 0.7 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.1 | 0.3 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.1 | 0.5 | GO:0046040 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.1 | GO:0002887 | positive regulation of cellular extravasation(GO:0002693) negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.1 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.8 | GO:0051318 | G1 phase(GO:0051318) |
| 0.1 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.6 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.3 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.4 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.3 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.3 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:1905207 | regulation of cardioblast differentiation(GO:0051890) regulation of cardiocyte differentiation(GO:1905207) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.0 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.0 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 1.1 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0071450 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:1904375 | regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of plasma membrane organization(GO:1903729) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.0 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.2 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 1.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0048703 | embryonic neurocranium morphogenesis(GO:0048702) embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.0 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.0 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1901998 | tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0002883 | hypersensitivity(GO:0002524) regulation of hypersensitivity(GO:0002883) positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 342.7 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0032153 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.7 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 1.2 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.4 | GO:0030427 | growth cone(GO:0030426) site of polarized growth(GO:0030427) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.4 | 2.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.3 | 1.6 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 1.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.3 | 0.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.5 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.3 | 2.6 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.2 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 0.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 346.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.2 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.6 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.1 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.2 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.8 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.4 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.0 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.0 | 2.5 | GO:0017171 | serine hydrolase activity(GO:0017171) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.0 | GO:0002060 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 1.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 191.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.2 | 2.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |