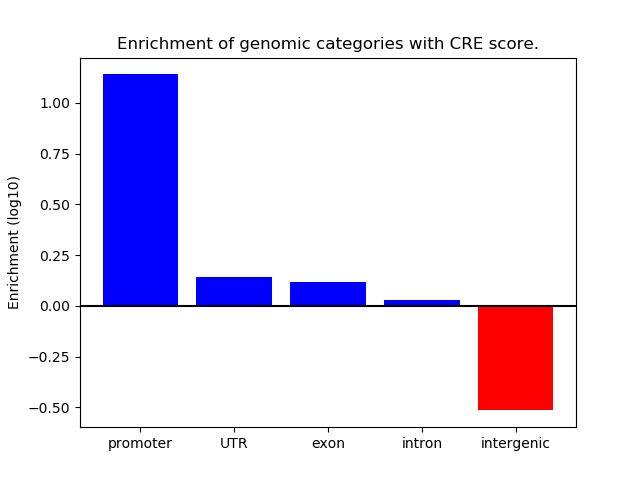
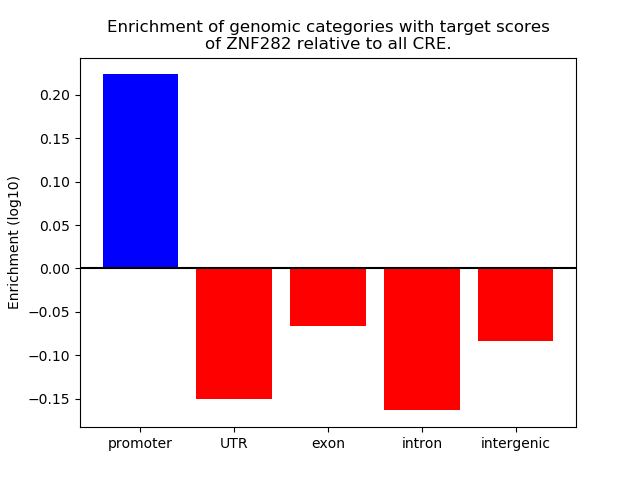
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
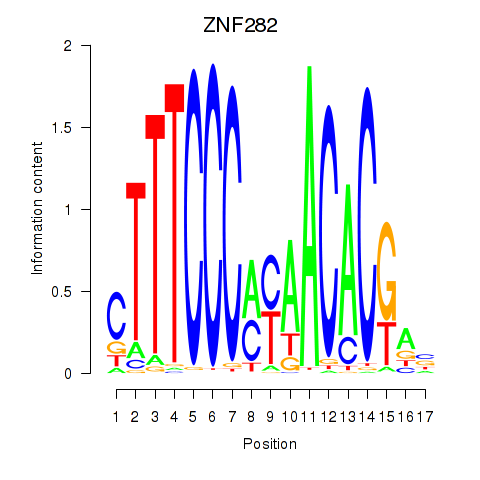
Results for ZNF282
Z-value: 1.42
Transcription factors associated with ZNF282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF282
|
ENSG00000170265.7 | zinc finger protein 282 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_148888070_148888331 | ZNF282 | 4377 | 0.181938 | 0.68 | 4.5e-02 | Click! |
| chr7_148878257_148878408 | ZNF282 | 14245 | 0.143314 | -0.67 | 4.6e-02 | Click! |
| chr7_148893161_148893454 | ZNF282 | 665 | 0.660005 | 0.63 | 6.9e-02 | Click! |
| chr7_148893506_148893991 | ZNF282 | 1106 | 0.456158 | 0.41 | 2.8e-01 | Click! |
| chr7_148888671_148888822 | ZNF282 | 3831 | 0.190840 | 0.40 | 2.8e-01 | Click! |
Activity of the ZNF282 motif across conditions
Conditions sorted by the z-value of the ZNF282 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
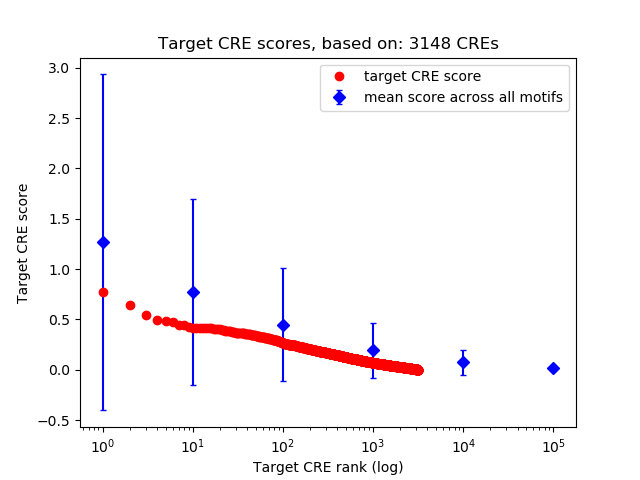
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_46572021_46572172 | 0.77 |
SLC38A1 |
solute carrier family 38, member 1 |
89388 |
0.1 |
| chr17_39969662_39969906 | 0.65 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
601 |
0.45 |
| chr11_119954135_119954286 | 0.55 |
TRIM29 |
tripartite motif containing 29 |
37398 |
0.19 |
| chr7_100771927_100772106 | 0.49 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
1637 |
0.23 |
| chr16_58534463_58534728 | 0.48 |
NDRG4 |
NDRG family member 4 |
545 |
0.68 |
| chr11_119953003_119953290 | 0.48 |
TRIM29 |
tripartite motif containing 29 |
38462 |
0.18 |
| chr7_1959519_1959814 | 0.45 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
20519 |
0.22 |
| chr5_125936367_125936518 | 0.44 |
PHAX |
phosphorylated adaptor for RNA export |
482 |
0.79 |
| chr1_88798905_88799056 | 0.42 |
ENSG00000239504 |
. |
144817 |
0.05 |
| chr5_67512014_67512283 | 0.42 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
544 |
0.82 |
| chr15_94544847_94544998 | 0.42 |
ENSG00000212022 |
. |
79498 |
0.11 |
| chr2_74780571_74780754 | 0.42 |
LOXL3 |
lysyl oxidase-like 3 |
400 |
0.5 |
| chr8_96307989_96308140 | 0.42 |
C8orf37 |
chromosome 8 open reading frame 37 |
26635 |
0.24 |
| chr11_18719924_18720222 | 0.41 |
TMEM86A |
transmembrane protein 86A |
265 |
0.89 |
| chr5_137688953_137689239 | 0.41 |
KDM3B |
lysine (K)-specific demethylase 3B |
811 |
0.51 |
| chr1_150971821_150972085 | 0.41 |
FAM63A |
family with sequence similarity 63, member A |
1307 |
0.24 |
| chr1_163038601_163039117 | 0.41 |
RGS4 |
regulator of G-protein signaling 4 |
76 |
0.98 |
| chr3_25471806_25472309 | 0.41 |
RARB |
retinoic acid receptor, beta |
2255 |
0.41 |
| chr6_10748064_10748484 | 0.41 |
TMEM14B |
transmembrane protein 14B |
210 |
0.53 |
| chr21_38711081_38711320 | 0.40 |
AP001437.1 |
|
27034 |
0.16 |
| chr8_55294341_55294557 | 0.39 |
ENSG00000244107 |
. |
38579 |
0.17 |
| chrX_100074585_100074736 | 0.39 |
CSTF2 |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
724 |
0.63 |
| chr20_50045192_50045343 | 0.39 |
ENSG00000266761 |
. |
24247 |
0.22 |
| chr1_226069091_226069419 | 0.38 |
TMEM63A |
transmembrane protein 63A |
814 |
0.51 |
| chr10_11865085_11865286 | 0.38 |
PROSER2 |
proline and serine-rich protein 2 |
212 |
0.95 |
| chr12_77719208_77719557 | 0.38 |
ENSG00000238769 |
. |
162577 |
0.04 |
| chr2_161491867_161492018 | 0.38 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
141637 |
0.05 |
| chrX_46432817_46433047 | 0.38 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
287 |
0.92 |
| chr2_157189525_157189762 | 0.37 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
411 |
0.9 |
| chr2_145276293_145276624 | 0.37 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
434 |
0.52 |
| chr10_114820084_114820658 | 0.37 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
83308 |
0.1 |
| chr11_119954548_119954699 | 0.37 |
TRIM29 |
tripartite motif containing 29 |
36985 |
0.19 |
| chr20_57586431_57586603 | 0.37 |
CTSZ |
cathepsin Z |
4215 |
0.17 |
| chr3_16006218_16006514 | 0.37 |
ENSG00000207815 |
. |
91088 |
0.08 |
| chr20_43150833_43151092 | 0.36 |
SERINC3 |
serine incorporator 3 |
212 |
0.91 |
| chr4_2486639_2486842 | 0.36 |
RNF4 |
ring finger protein 4 |
4798 |
0.2 |
| chr7_149572287_149572438 | 0.36 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
1230 |
0.35 |
| chr1_197887720_197887871 | 0.36 |
LHX9 |
LIM homeobox 9 |
1266 |
0.53 |
| chr17_7121037_7121301 | 0.36 |
DLG4 |
discs, large homolog 4 (Drosophila) |
503 |
0.43 |
| chr12_125412025_125412330 | 0.36 |
UBC |
ubiquitin C |
10263 |
0.16 |
| chr12_127256953_127257104 | 0.35 |
ENSG00000239776 |
. |
393644 |
0.01 |
| chr9_96930088_96930349 | 0.35 |
ENSG00000199165 |
. |
8016 |
0.16 |
| chr6_165722376_165722527 | 0.35 |
C6orf118 |
chromosome 6 open reading frame 118 |
109 |
0.98 |
| chr15_75471507_75471789 | 0.35 |
C15orf39 |
chromosome 15 open reading frame 39 |
16336 |
0.13 |
| chr1_161369629_161370065 | 0.35 |
ENSG00000206921 |
. |
1232 |
0.42 |
| chr19_13977402_13977553 | 0.34 |
ENSG00000207613 |
. |
8036 |
0.08 |
| chr2_225846726_225846877 | 0.34 |
ENSG00000263828 |
. |
28456 |
0.22 |
| chr14_63672700_63672938 | 0.34 |
RHOJ |
ras homolog family member J |
1242 |
0.54 |
| chr20_21961725_21961876 | 0.34 |
PAX1 |
paired box 1 |
274939 |
0.02 |
| chrX_65220606_65220757 | 0.34 |
RP6-159A1.3 |
|
1088 |
0.56 |
| chr21_42739358_42739509 | 0.33 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
2579 |
0.29 |
| chr1_243646920_243647071 | 0.33 |
RP11-269F20.1 |
|
61839 |
0.14 |
| chr6_34358484_34358635 | 0.33 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
1892 |
0.33 |
| chr5_58335725_58336097 | 0.33 |
RP11-266N13.2 |
|
323 |
0.69 |
| chr8_13372824_13372975 | 0.33 |
DLC1 |
deleted in liver cancer 1 |
503 |
0.82 |
| chr12_65672666_65673070 | 0.33 |
MSRB3 |
methionine sulfoxide reductase B3 |
101 |
0.97 |
| chr7_93943609_93943760 | 0.33 |
COL1A2 |
collagen, type I, alpha 2 |
80189 |
0.11 |
| chr7_156687876_156688027 | 0.33 |
LMBR1 |
limb development membrane protein 1 |
2027 |
0.34 |
| chr9_116356768_116357019 | 0.33 |
RGS3 |
regulator of G-protein signaling 3 |
1127 |
0.54 |
| chrX_100183665_100184190 | 0.33 |
XKRX |
XK, Kell blood group complex subunit-related, X-linked |
29 |
0.98 |
| chr17_61722686_61722837 | 0.32 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
22691 |
0.13 |
| chrX_133687659_133687810 | 0.32 |
ENSG00000223749 |
. |
6993 |
0.12 |
| chr20_8651278_8651429 | 0.32 |
ENSG00000222298 |
. |
60466 |
0.12 |
| chr5_25569497_25569648 | 0.31 |
ENSG00000201016 |
. |
131861 |
0.06 |
| chr2_242319557_242319708 | 0.31 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
7368 |
0.16 |
| chr1_87223134_87223385 | 0.31 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
52681 |
0.12 |
| chr1_226852545_226852696 | 0.31 |
ITPKB-AS1 |
ITPKB antisense RNA 1 |
3978 |
0.24 |
| chr12_2420181_2420332 | 0.31 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
41314 |
0.18 |
| chr17_37844389_37844572 | 0.31 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
87 |
0.53 |
| chr16_1979223_1979447 | 0.31 |
HS3ST6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
10894 |
0.06 |
| chr11_82774150_82774358 | 0.31 |
RAB30 |
RAB30, member RAS oncogene family |
8616 |
0.18 |
| chrY_6883288_6883439 | 0.31 |
TBL1Y |
transducin (beta)-like 1, Y-linked |
104636 |
0.08 |
| chr10_114206747_114206976 | 0.31 |
VTI1A |
vesicle transport through interaction with t-SNAREs 1A |
155 |
0.54 |
| chr15_41850580_41851004 | 0.31 |
TYRO3 |
TYRO3 protein tyrosine kinase |
440 |
0.79 |
| chr12_113841803_113841954 | 0.31 |
SDS |
serine dehydratase |
185 |
0.94 |
| chrX_46616562_46616983 | 0.30 |
SLC9A7 |
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
1718 |
0.46 |
| chr19_13319063_13319297 | 0.30 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
49034 |
0.08 |
| chr8_97170599_97170750 | 0.30 |
GDF6 |
growth differentiation factor 6 |
2346 |
0.36 |
| chr1_221342999_221343150 | 0.30 |
HLX |
H2.0-like homeobox |
288490 |
0.01 |
| chr10_89265401_89265555 | 0.30 |
MINPP1 |
multiple inositol-polyphosphate phosphatase 1 |
846 |
0.69 |
| chrY_19466305_19466456 | 0.30 |
ENSG00000252315 |
. |
203364 |
0.03 |
| chr1_183605727_183605878 | 0.30 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
558 |
0.58 |
| chr8_23571667_23572036 | 0.29 |
RP11-175E9.1 |
|
7197 |
0.17 |
| chr11_66444702_66445046 | 0.29 |
RBM4B |
RNA binding motif protein 4B |
345 |
0.79 |
| chr20_2451568_2451719 | 0.29 |
SNRPB |
small nuclear ribonucleoprotein polypeptides B and B1 |
144 |
0.94 |
| chr15_62353285_62353511 | 0.29 |
RP11-643M14.1 |
|
685 |
0.44 |
| chr15_40234807_40235047 | 0.29 |
ENSG00000238564 |
. |
7980 |
0.15 |
| chr11_119954900_119955051 | 0.28 |
TRIM29 |
tripartite motif containing 29 |
36633 |
0.19 |
| chr19_6110825_6111002 | 0.28 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
301 |
0.84 |
| chr8_37344900_37345051 | 0.28 |
RP11-150O12.6 |
|
29564 |
0.23 |
| chr17_46627635_46627871 | 0.28 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
761 |
0.38 |
| chr1_86043929_86044119 | 0.28 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
91 |
0.97 |
| chr3_123477356_123477507 | 0.28 |
MYLK |
myosin light chain kinase |
35257 |
0.15 |
| chr14_61447182_61447373 | 0.28 |
TRMT5 |
tRNA methyltransferase 5 |
492 |
0.5 |
| chr17_63341710_63341861 | 0.28 |
ENSG00000265189 |
. |
74993 |
0.12 |
| chr1_221910890_221911087 | 0.28 |
DUSP10 |
dual specificity phosphatase 10 |
186 |
0.97 |
| chr6_24667419_24667634 | 0.28 |
ACOT13 |
acyl-CoA thioesterase 13 |
227 |
0.53 |
| chr5_128429913_128430155 | 0.27 |
ISOC1 |
isochorismatase domain containing 1 |
410 |
0.87 |
| chr20_31988038_31988189 | 0.27 |
CDK5RAP1 |
CDK5 regulatory subunit associated protein 1 |
1210 |
0.46 |
| chr5_5419342_5419575 | 0.27 |
KIAA0947 |
KIAA0947 |
3349 |
0.39 |
| chr17_47077722_47077873 | 0.27 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
2689 |
0.14 |
| chr14_58862127_58862433 | 0.26 |
RP11-517O13.3 |
|
79 |
0.8 |
| chr11_72449633_72449911 | 0.26 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11599 |
0.12 |
| chr9_107755904_107756060 | 0.26 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
65464 |
0.11 |
| chr7_101930639_101930983 | 0.26 |
SH2B2 |
SH2B adaptor protein 2 |
2359 |
0.2 |
| chr10_53459424_53459629 | 0.26 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
171 |
0.97 |
| chr11_118939891_118940176 | 0.25 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
1540 |
0.15 |
| chr15_93446443_93446658 | 0.25 |
ENSG00000264173 |
. |
1079 |
0.38 |
| chr16_22202801_22203159 | 0.25 |
EEF2K |
eukaryotic elongation factor-2 kinase |
14623 |
0.17 |
| chr6_26313507_26313734 | 0.25 |
HIST1H4H |
histone cluster 1, H4h |
27858 |
0.07 |
| chr7_89896255_89896406 | 0.25 |
AC002064.4 |
|
3200 |
0.19 |
| chr6_133133816_133133967 | 0.25 |
RPS12 |
ribosomal protein S12 |
1689 |
0.22 |
| chr13_98112435_98112620 | 0.25 |
RAP2A |
RAP2A, member of RAS oncogene family |
26051 |
0.24 |
| chr8_32559587_32559738 | 0.25 |
NRG1 |
neuregulin 1 |
19609 |
0.29 |
| chr18_71857391_71857542 | 0.25 |
TIMM21 |
translocase of inner mitochondrial membrane 21 homolog (yeast) |
35056 |
0.15 |
| chr7_131338473_131338624 | 0.25 |
PODXL |
podocalyxin-like |
97172 |
0.08 |
| chr4_81189028_81189189 | 0.25 |
FGF5 |
fibroblast growth factor 5 |
1315 |
0.54 |
| chr1_201857476_201857627 | 0.25 |
SHISA4 |
shisa family member 4 |
285 |
0.85 |
| chr1_90460194_90460489 | 0.25 |
ZNF326 |
zinc finger protein 326 |
333 |
0.91 |
| chr11_119953470_119953621 | 0.25 |
TRIM29 |
tripartite motif containing 29 |
38063 |
0.18 |
| chr1_109504839_109504990 | 0.25 |
CLCC1 |
chloride channel CLIC-like 1 |
1158 |
0.34 |
| chr12_52346063_52346405 | 0.25 |
ACVR1B |
activin A receptor, type IB |
706 |
0.62 |
| chr5_122110815_122111189 | 0.25 |
SNX2 |
sorting nexin 2 |
104 |
0.94 |
| chr8_27630129_27630324 | 0.25 |
CCDC25 |
coiled-coil domain containing 25 |
69 |
0.89 |
| chr5_172185603_172185754 | 0.25 |
RP11-779O18.1 |
|
449 |
0.76 |
| chrX_135848946_135849996 | 0.25 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
31 |
0.98 |
| chr1_240256340_240256520 | 0.25 |
FMN2 |
formin 2 |
1250 |
0.57 |
| chr9_125796489_125796713 | 0.25 |
GPR21 |
G protein-coupled receptor 21 |
205 |
0.86 |
| chr12_53846681_53847472 | 0.25 |
PCBP2 |
poly(rC) binding protein 2 |
448 |
0.72 |
| chr1_89591012_89591781 | 0.25 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
401 |
0.85 |
| chr22_40571850_40572001 | 0.24 |
TNRC6B |
trinucleotide repeat containing 6B |
2004 |
0.43 |
| chr15_79328270_79328421 | 0.24 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
30338 |
0.17 |
| chr16_66586477_66586958 | 0.24 |
CKLF-CMTM1 |
CKLF-CMTM1 readthrough |
165 |
0.44 |
| chr6_146284922_146285161 | 0.24 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
192 |
0.97 |
| chr19_55765035_55765186 | 0.24 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
2027 |
0.15 |
| chr17_18313829_18314117 | 0.24 |
AL353997.4 |
|
10160 |
0.11 |
| chr20_4954300_4954516 | 0.24 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
27737 |
0.17 |
| chrX_129065192_129065438 | 0.24 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
17976 |
0.18 |
| chr4_140375324_140375815 | 0.24 |
RP11-83A24.2 |
|
381 |
0.62 |
| chr1_170640082_170640341 | 0.24 |
PRRX1 |
paired related homeobox 1 |
7133 |
0.3 |
| chr2_162849046_162849197 | 0.24 |
ENSG00000253046 |
. |
73829 |
0.1 |
| chr7_114459536_114459687 | 0.24 |
MDFIC |
MyoD family inhibitor domain containing |
102598 |
0.08 |
| chr2_177042560_177043156 | 0.24 |
HOXD-AS1 |
HOXD cluster antisense RNA 1 |
5 |
0.95 |
| chr10_31608301_31608721 | 0.24 |
ENSG00000237036 |
. |
109 |
0.8 |
| chr15_60898079_60898454 | 0.23 |
RORA |
RAR-related orphan receptor A |
12941 |
0.23 |
| chr5_154078594_154078745 | 0.23 |
ENSG00000263478 |
. |
4675 |
0.17 |
| chr11_85358415_85358894 | 0.23 |
TMEM126A |
transmembrane protein 126A |
367 |
0.83 |
| chr12_68043814_68044335 | 0.23 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
612 |
0.81 |
| chr2_55647724_55647875 | 0.23 |
CCDC88A |
coiled-coil domain containing 88A |
742 |
0.62 |
| chrX_102631036_102631748 | 0.23 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
57 |
0.97 |
| chr7_82201510_82201661 | 0.23 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
128471 |
0.06 |
| chr9_119713843_119714110 | 0.23 |
ENSG00000265662 |
. |
147310 |
0.04 |
| chr19_10811895_10812062 | 0.23 |
QTRT1 |
queuine tRNA-ribosyltransferase 1 |
149 |
0.93 |
| chr13_99160815_99160995 | 0.23 |
STK24 |
serine/threonine kinase 24 |
10616 |
0.23 |
| chr20_12990547_12990774 | 0.23 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
663 |
0.83 |
| chr15_64679591_64679870 | 0.23 |
TRIP4 |
thyroid hormone receptor interactor 4 |
273 |
0.85 |
| chr4_1241678_1241899 | 0.23 |
CTBP1 |
C-terminal binding protein 1 |
740 |
0.5 |
| chr20_9073959_9074110 | 0.23 |
PLCB4 |
phospholipase C, beta 4 |
24286 |
0.22 |
| chrX_87155513_87155664 | 0.23 |
ENSG00000252016 |
. |
248205 |
0.02 |
| chr9_133712337_133712924 | 0.23 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
2177 |
0.35 |
| chr1_67600256_67600407 | 0.23 |
C1orf141 |
chromosome 1 open reading frame 141 |
308 |
0.89 |
| chr12_133404529_133404680 | 0.22 |
GOLGA3 |
golgin A3 |
684 |
0.62 |
| chr18_20274804_20275027 | 0.22 |
RP11-370A5.1 |
|
36613 |
0.17 |
| chr6_132179827_132179978 | 0.22 |
ENSG00000200895 |
. |
38428 |
0.14 |
| chr1_206645642_206645925 | 0.22 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
1945 |
0.27 |
| chr3_155393866_155394017 | 0.22 |
PLCH1 |
phospholipase C, eta 1 |
113 |
0.98 |
| chr3_16927344_16927610 | 0.22 |
PLCL2 |
phospholipase C-like 2 |
1025 |
0.65 |
| chr19_16461941_16462107 | 0.22 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
10740 |
0.14 |
| chr20_48955472_48955638 | 0.22 |
ENSG00000244376 |
. |
90465 |
0.08 |
| chr1_163845312_163845463 | 0.22 |
ENSG00000212538 |
. |
47520 |
0.2 |
| chr2_175546375_175546968 | 0.22 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
928 |
0.66 |
| chr12_49450678_49450829 | 0.22 |
KMT2D |
lysine (K)-specific methyltransferase 2D |
1646 |
0.2 |
| chr12_125473806_125474002 | 0.22 |
DHX37 |
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
237 |
0.93 |
| chr1_12139606_12139757 | 0.22 |
ENSG00000201135 |
. |
1640 |
0.3 |
| chr14_103481209_103481581 | 0.22 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
42404 |
0.13 |
| chr8_75234014_75234228 | 0.22 |
JPH1 |
junctophilin 1 |
558 |
0.76 |
| chr5_175788940_175789140 | 0.22 |
KIAA1191 |
KIAA1191 |
69 |
0.94 |
| chr15_34627792_34628213 | 0.21 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
1043 |
0.38 |
| chr20_13619088_13619556 | 0.21 |
TASP1 |
taspase, threonine aspartase, 1 |
228 |
0.96 |
| chr8_124829666_124829817 | 0.21 |
FER1L6 |
fer-1-like 6 (C. elegans) |
34486 |
0.17 |
| chr11_111957062_111957410 | 0.21 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
261 |
0.49 |
| chr3_125931550_125931701 | 0.21 |
ALDH1L1-AS2 |
ALDH1L1 antisense RNA 2 |
5527 |
0.19 |
| chr5_14983876_14984027 | 0.21 |
ENSG00000212036 |
. |
73202 |
0.11 |
| chr3_177624174_177624369 | 0.21 |
ENSG00000199858 |
. |
101296 |
0.09 |
| chrX_12992120_12992286 | 0.21 |
TMSB4X |
thymosin beta 4, X-linked |
1024 |
0.6 |
| chrX_118924681_118925547 | 0.21 |
RPL39 |
ribosomal protein L39 |
492 |
0.74 |
| chr2_216286742_216286974 | 0.21 |
FN1 |
fibronectin 1 |
13932 |
0.21 |
| chr2_241497193_241497674 | 0.21 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
5 |
0.96 |
| chr21_35748199_35748651 | 0.21 |
SMIM11 |
small integral membrane protein 11 |
522 |
0.54 |
| chr5_168167573_168167764 | 0.21 |
ENSG00000207739 |
. |
27592 |
0.17 |
| chr12_47219516_47219750 | 0.21 |
SLC38A4 |
solute carrier family 38, member 4 |
135 |
0.98 |
| chr18_60520045_60520196 | 0.21 |
AC015989.2 |
|
14172 |
0.2 |
| chr4_119201779_119201930 | 0.21 |
ENSG00000269893 |
. |
1509 |
0.54 |
| chr7_143083179_143083339 | 0.21 |
ZYX |
zyxin |
3166 |
0.14 |
| chr15_83420399_83420550 | 0.21 |
RP11-752G15.6 |
|
1138 |
0.35 |
| chr19_47731656_47732088 | 0.20 |
ENSG00000265134 |
. |
1673 |
0.27 |
| chr18_72162856_72163393 | 0.20 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
73 |
0.98 |
| chr4_144434579_144434842 | 0.20 |
SMARCA5 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
94 |
0.95 |
| chr11_65480098_65480381 | 0.20 |
KAT5 |
K(lysine) acetyltransferase 5 |
37 |
0.95 |
| chr1_149899868_149900298 | 0.20 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
153 |
0.9 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.4 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.4 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.1 | 0.2 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.2 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.1 | 0.2 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0010664 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.4 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.2 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.2 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.3 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.2 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.2 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.4 | GO:0044766 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0042987 | amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.1 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.3 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.1 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |