Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
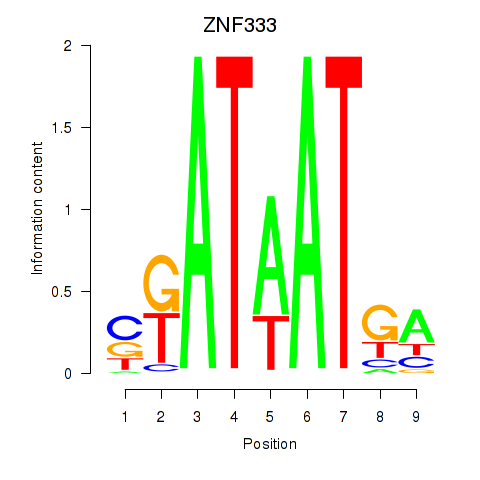
Results for ZNF333
Z-value: 2.40
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
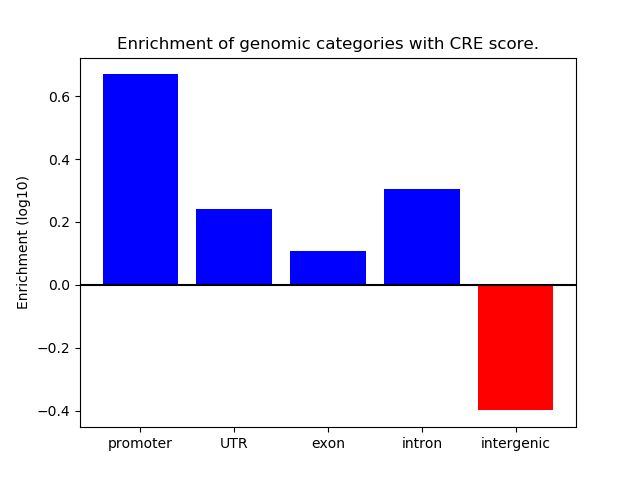
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_14793991_14794142 | ZNF333 | 6645 | 0.148571 | -0.63 | 6.8e-02 | Click! |
| chr19_14801111_14801506 | ZNF333 | 395 | 0.514246 | -0.49 | 1.8e-01 | Click! |
| chr19_14800223_14800431 | ZNF333 | 384 | 0.555379 | -0.44 | 2.4e-01 | Click! |
| chr19_14800609_14800883 | ZNF333 | 35 | 0.558616 | -0.39 | 3.0e-01 | Click! |
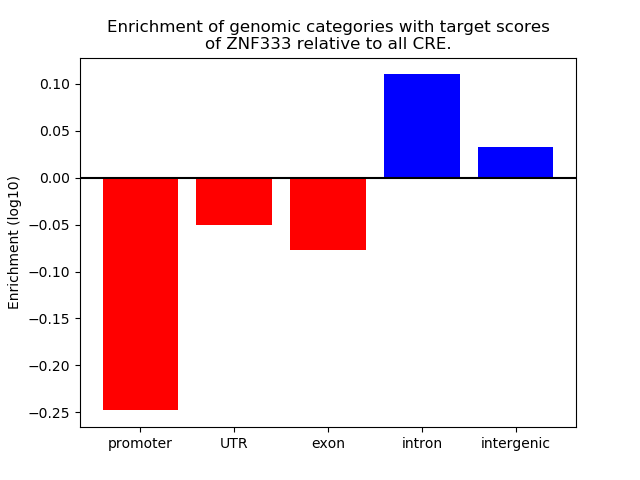
Activity of the ZNF333 motif across conditions
Conditions sorted by the z-value of the ZNF333 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_40301249_40301400 | 0.66 |
GRAP2 |
GRB2-related adaptor protein 2 |
4211 |
0.2 |
| chr1_84612029_84612357 | 0.60 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
2239 |
0.42 |
| chr7_38312198_38312349 | 0.58 |
STARD3NL |
STARD3 N-terminal like |
94276 |
0.09 |
| chr10_17705373_17705524 | 0.53 |
ENSG00000251959 |
. |
15739 |
0.14 |
| chr10_25176560_25176711 | 0.52 |
ENSG00000240294 |
. |
21106 |
0.23 |
| chr8_93110125_93110276 | 0.51 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
1684 |
0.54 |
| chr11_118210860_118211247 | 0.51 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
599 |
0.61 |
| chr8_58961464_58961615 | 0.50 |
FAM110B |
family with sequence similarity 110, member B |
54426 |
0.18 |
| chrX_135730916_135731129 | 0.50 |
CD40LG |
CD40 ligand |
636 |
0.7 |
| chr7_142511951_142512102 | 0.48 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
30895 |
0.13 |
| chr5_109028785_109028975 | 0.47 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
3813 |
0.23 |
| chr12_21606326_21606605 | 0.46 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
15616 |
0.17 |
| chr14_61802494_61802645 | 0.46 |
PRKCH |
protein kinase C, eta |
8391 |
0.2 |
| chr5_158520160_158520311 | 0.45 |
EBF1 |
early B-cell factor 1 |
6466 |
0.27 |
| chr3_86894495_86894646 | 0.45 |
VGLL3 |
vestigial like 3 (Drosophila) |
145282 |
0.05 |
| chr6_90790442_90790643 | 0.45 |
ENSG00000222078 |
. |
79317 |
0.1 |
| chr2_204734743_204734894 | 0.44 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
137 |
0.97 |
| chr4_143321559_143321710 | 0.44 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30778 |
0.27 |
| chr7_38400564_38400715 | 0.43 |
AMPH |
amphiphysin |
102074 |
0.08 |
| chr14_100531999_100532513 | 0.42 |
EVL |
Enah/Vasp-like |
498 |
0.76 |
| chr3_186743627_186743921 | 0.41 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
503 |
0.85 |
| chr1_160489868_160490211 | 0.41 |
SLAMF6 |
SLAM family member 6 |
2993 |
0.18 |
| chr1_207093221_207093431 | 0.41 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
1886 |
0.27 |
| chr13_50028881_50029032 | 0.41 |
AL136218.1 |
Sarcoma antigen NY-SAR-79; Uncharacterized protein |
8402 |
0.17 |
| chr14_55769874_55770025 | 0.40 |
FBXO34 |
F-box protein 34 |
31077 |
0.17 |
| chr11_118176160_118176414 | 0.40 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
673 |
0.59 |
| chr6_90791046_90791197 | 0.40 |
ENSG00000222078 |
. |
79896 |
0.1 |
| chr11_97853847_97853998 | 0.40 |
ENSG00000216073 |
. |
70033 |
0.14 |
| chr13_99955816_99955967 | 0.40 |
GPR183 |
G protein-coupled receptor 183 |
3768 |
0.25 |
| chr5_159897818_159898005 | 0.40 |
ENSG00000265237 |
. |
3498 |
0.21 |
| chr4_109086470_109087224 | 0.40 |
LEF1 |
lymphoid enhancer-binding factor 1 |
610 |
0.72 |
| chr14_99702706_99702901 | 0.40 |
AL109767.1 |
|
26482 |
0.19 |
| chr9_33162104_33162255 | 0.40 |
RP11-326F20.5 |
|
4794 |
0.16 |
| chr5_85693756_85693907 | 0.39 |
NBPF22P |
neuroblastoma breakpoint family, member 22, pseudogene |
115247 |
0.07 |
| chr4_20289069_20289220 | 0.38 |
SLIT2 |
slit homolog 2 (Drosophila) |
32601 |
0.24 |
| chr1_207092040_207092191 | 0.38 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
3097 |
0.19 |
| chr13_99909728_99909879 | 0.38 |
GPR18 |
G protein-coupled receptor 18 |
825 |
0.64 |
| chr8_82051121_82051272 | 0.38 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
26893 |
0.24 |
| chr1_111212431_111212582 | 0.37 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
5149 |
0.2 |
| chr7_107386589_107386740 | 0.37 |
AC002467.7 |
|
1638 |
0.29 |
| chr4_83930221_83930372 | 0.37 |
LIN54 |
lin-54 homolog (C. elegans) |
1566 |
0.33 |
| chr22_20905629_20905785 | 0.37 |
MED15 |
mediator complex subunit 15 |
130 |
0.95 |
| chr14_22955963_22956254 | 0.37 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
63 |
0.95 |
| chr19_2892956_2893107 | 0.37 |
ZNF57 |
zinc finger protein 57 |
7865 |
0.11 |
| chr12_92440588_92440837 | 0.37 |
C12orf79 |
chromosome 12 open reading frame 79 |
90085 |
0.08 |
| chrX_110366293_110366528 | 0.37 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
105 |
0.98 |
| chr10_33239572_33239908 | 0.36 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
4851 |
0.32 |
| chrX_103515232_103515731 | 0.36 |
ESX1 |
ESX homeobox 1 |
15867 |
0.25 |
| chr1_193319454_193319605 | 0.36 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
163745 |
0.04 |
| chr5_19885020_19885171 | 0.36 |
CDH18 |
cadherin 18, type 2 |
1286 |
0.64 |
| chr10_69231096_69231247 | 0.36 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
176100 |
0.03 |
| chr7_38392814_38392965 | 0.36 |
AMPH |
amphiphysin |
109824 |
0.07 |
| chr13_99949671_99949957 | 0.35 |
GPR183 |
G protein-coupled receptor 183 |
9845 |
0.2 |
| chr3_68107536_68107743 | 0.35 |
FAM19A1 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
52266 |
0.19 |
| chr11_94802718_94802918 | 0.35 |
ENDOD1 |
endonuclease domain containing 1 |
20156 |
0.19 |
| chr21_32558355_32558506 | 0.35 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
55891 |
0.16 |
| chr2_177679359_177679510 | 0.35 |
ENSG00000206866 |
. |
115384 |
0.06 |
| chr2_235401897_235402222 | 0.35 |
ARL4C |
ADP-ribosylation factor-like 4C |
3185 |
0.41 |
| chr3_150686052_150686203 | 0.35 |
CLRN1-AS1 |
CLRN1 antisense RNA 1 |
4338 |
0.2 |
| chr7_28741672_28741823 | 0.34 |
CREB5 |
cAMP responsive element binding protein 5 |
16149 |
0.3 |
| chr3_12332222_12332373 | 0.34 |
PPARG |
peroxisome proliferator-activated receptor gamma |
1727 |
0.49 |
| chr1_86848612_86848888 | 0.34 |
ODF2L |
outer dense fiber of sperm tails 2-like |
129 |
0.97 |
| chr2_237077720_237077871 | 0.34 |
GBX2 |
gastrulation brain homeobox 2 |
783 |
0.52 |
| chr5_74347596_74347880 | 0.34 |
GCNT4 |
glucosaminyl (N-acetyl) transferase 4, core 2 |
21014 |
0.25 |
| chr2_181995547_181995698 | 0.34 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
148872 |
0.04 |
| chr3_195806787_195806938 | 0.34 |
TFRC |
transferrin receptor |
2099 |
0.29 |
| chr21_19166563_19166714 | 0.33 |
AL109761.5 |
|
833 |
0.67 |
| chr12_47607489_47607701 | 0.33 |
PCED1B |
PC-esterase domain containing 1B |
2457 |
0.32 |
| chr14_41685035_41685186 | 0.33 |
ENSG00000221695 |
. |
133387 |
0.06 |
| chr3_158393631_158393782 | 0.33 |
LXN |
latexin |
3224 |
0.22 |
| chr11_129716903_129717070 | 0.33 |
TMEM45B |
transmembrane protein 45B |
3241 |
0.32 |
| chr9_14180845_14181011 | 0.33 |
NFIB |
nuclear factor I/B |
131 |
0.98 |
| chr1_209998785_209998936 | 0.33 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
2492 |
0.24 |
| chr11_128378341_128378619 | 0.33 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
3191 |
0.31 |
| chr13_31348514_31348665 | 0.33 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
38944 |
0.19 |
| chrX_57931578_57932092 | 0.33 |
ZXDA |
zinc finger, X-linked, duplicated A |
5232 |
0.36 |
| chr2_136876365_136876597 | 0.33 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
746 |
0.77 |
| chr19_42056193_42056486 | 0.33 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
453 |
0.8 |
| chr22_40297106_40297391 | 0.32 |
GRAP2 |
GRB2-related adaptor protein 2 |
135 |
0.96 |
| chr11_57494574_57494725 | 0.32 |
C11orf31 |
chromosome 11 open reading frame 31 |
14176 |
0.1 |
| chr5_39197523_39197702 | 0.32 |
FYB |
FYN binding protein |
5517 |
0.3 |
| chr14_41187793_41187944 | 0.32 |
ENSG00000221695 |
. |
630629 |
0.0 |
| chr13_99953472_99953749 | 0.32 |
GPR183 |
G protein-coupled receptor 183 |
6049 |
0.22 |
| chr2_204801338_204802214 | 0.32 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr3_151936530_151936714 | 0.32 |
MBNL1 |
muscleblind-like splicing regulator 1 |
49207 |
0.15 |
| chr12_46603502_46603653 | 0.32 |
SLC38A1 |
solute carrier family 38, member 1 |
57907 |
0.16 |
| chr6_63090511_63090662 | 0.32 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
94454 |
0.1 |
| chr18_74837172_74837323 | 0.32 |
MBP |
myelin basic protein |
2421 |
0.42 |
| chr12_56329135_56329873 | 0.32 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
114 |
0.92 |
| chr1_117302165_117302316 | 0.32 |
CD2 |
CD2 molecule |
5151 |
0.24 |
| chr2_197047793_197047944 | 0.32 |
STK17B |
serine/threonine kinase 17b |
6641 |
0.2 |
| chr6_128229281_128229524 | 0.32 |
THEMIS |
thymocyte selection associated |
7176 |
0.3 |
| chr1_101705180_101705331 | 0.31 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
2811 |
0.2 |
| chr6_143178538_143178701 | 0.31 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
20435 |
0.26 |
| chr3_128207705_128209147 | 0.31 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr2_208108634_208108785 | 0.31 |
AC007879.5 |
|
10267 |
0.22 |
| chr10_62487366_62487517 | 0.31 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
5807 |
0.3 |
| chr17_56654648_56654799 | 0.31 |
C17orf47 |
chromosome 17 open reading frame 47 |
32994 |
0.1 |
| chr1_246812163_246812314 | 0.31 |
RP11-439E19.1 |
|
43063 |
0.14 |
| chr7_130632583_130632734 | 0.31 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
36210 |
0.19 |
| chr5_156608214_156608444 | 0.31 |
ITK |
IL2-inducible T-cell kinase |
492 |
0.72 |
| chr8_97350508_97350718 | 0.31 |
ENSG00000199732 |
. |
29063 |
0.17 |
| chr3_50644588_50644940 | 0.31 |
CISH |
cytokine inducible SH2-containing protein |
4439 |
0.16 |
| chr7_103577846_103577997 | 0.31 |
RELN |
reelin |
52042 |
0.18 |
| chr4_16877414_16877627 | 0.31 |
LDB2 |
LIM domain binding 2 |
22665 |
0.29 |
| chr12_55374952_55375103 | 0.31 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
595 |
0.79 |
| chr4_40209794_40209945 | 0.31 |
RHOH |
ras homolog family member H |
7905 |
0.22 |
| chr6_122931066_122932065 | 0.30 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
188 |
0.97 |
| chr7_150146102_150146542 | 0.30 |
GIMAP8 |
GTPase, IMAP family member 8 |
1396 |
0.39 |
| chr17_266453_266873 | 0.30 |
AC108004.3 |
|
2849 |
0.2 |
| chr5_39186524_39186686 | 0.30 |
FYB |
FYN binding protein |
16524 |
0.26 |
| chr13_99958437_99958588 | 0.30 |
GPR183 |
G protein-coupled receptor 183 |
1147 |
0.52 |
| chr16_60708046_60708197 | 0.30 |
ENSG00000265127 |
. |
95762 |
0.1 |
| chr12_72523140_72523291 | 0.30 |
RP11-89M22.3 |
|
72058 |
0.12 |
| chr2_200470649_200470800 | 0.30 |
SATB2 |
SATB homeobox 2 |
134735 |
0.05 |
| chr6_89584193_89584392 | 0.30 |
RNGTT |
RNA guanylyltransferase and 5'-phosphatase |
88988 |
0.08 |
| chr20_43347005_43347156 | 0.30 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
3194 |
0.18 |
| chr3_87484012_87484163 | 0.30 |
POU1F1 |
POU class 1 homeobox 1 |
158350 |
0.04 |
| chr20_51591815_51591966 | 0.30 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
2944 |
0.37 |
| chr7_15730839_15730990 | 0.30 |
MEOX2 |
mesenchyme homeobox 2 |
4477 |
0.29 |
| chr19_6926549_6926700 | 0.30 |
RP11-1137G4.3 |
|
26598 |
0.12 |
| chr17_77001373_77001524 | 0.29 |
CANT1 |
calcium activated nucleotidase 1 |
4364 |
0.16 |
| chr12_12712693_12713134 | 0.29 |
DUSP16 |
dual specificity phosphatase 16 |
1098 |
0.58 |
| chr1_12445934_12446085 | 0.29 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
23915 |
0.22 |
| chr6_65662806_65662957 | 0.29 |
ENSG00000221312 |
. |
21405 |
0.3 |
| chr21_21342429_21342580 | 0.29 |
ENSG00000251851 |
. |
385660 |
0.01 |
| chr11_120195755_120196855 | 0.29 |
TMEM136 |
transmembrane protein 136 |
289 |
0.9 |
| chr3_98280847_98280998 | 0.29 |
GPR15 |
G protein-coupled receptor 15 |
30179 |
0.12 |
| chr11_46581548_46581763 | 0.29 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
31259 |
0.15 |
| chr5_64310480_64310631 | 0.29 |
ENSG00000207439 |
. |
108641 |
0.07 |
| chr20_55204347_55205219 | 0.29 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr7_114566372_114566651 | 0.29 |
MDFIC |
MyoD family inhibitor domain containing |
3380 |
0.39 |
| chr4_66185713_66185864 | 0.29 |
EPHA5 |
EPH receptor A5 |
349865 |
0.01 |
| chr2_237446083_237446280 | 0.28 |
IQCA1 |
IQ motif containing with AAA domain 1 |
29996 |
0.19 |
| chr14_22322504_22322655 | 0.28 |
ENSG00000222776 |
. |
73794 |
0.11 |
| chr6_43351299_43351450 | 0.28 |
ZNF318 |
zinc finger protein 318 |
14193 |
0.14 |
| chr12_119615001_119615152 | 0.28 |
RP11-64B16.3 |
|
787 |
0.51 |
| chr4_24732106_24732257 | 0.28 |
SOD3 |
superoxide dismutase 3, extracellular |
59353 |
0.15 |
| chr14_99735608_99735969 | 0.28 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
1777 |
0.38 |
| chr8_56777196_56777347 | 0.28 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
15101 |
0.15 |
| chr14_61790070_61790226 | 0.28 |
ENSG00000247287 |
. |
612 |
0.51 |
| chr10_14920855_14921866 | 0.28 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
439 |
0.82 |
| chr4_40210765_40210916 | 0.28 |
RHOH |
ras homolog family member H |
8876 |
0.22 |
| chr4_154410997_154411265 | 0.28 |
KIAA0922 |
KIAA0922 |
23630 |
0.22 |
| chr3_66940641_66940792 | 0.28 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
108011 |
0.08 |
| chr7_19139525_19139676 | 0.28 |
AC003986.6 |
|
12497 |
0.16 |
| chr12_40448353_40448504 | 0.28 |
SLC2A13 |
solute carrier family 2 (facilitated glucose transporter), member 13 |
51233 |
0.17 |
| chr6_119398487_119399078 | 0.28 |
FAM184A |
family with sequence similarity 184, member A |
755 |
0.67 |
| chr2_196444875_196445026 | 0.28 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
4249 |
0.31 |
| chr1_195164944_195165095 | 0.28 |
ENSG00000265108 |
. |
69208 |
0.15 |
| chr2_181570738_181570889 | 0.28 |
ENSG00000264976 |
. |
88880 |
0.1 |
| chr1_51171312_51171463 | 0.28 |
ENSG00000252825 |
. |
44581 |
0.17 |
| chr3_130176422_130176573 | 0.28 |
COL6A5 |
collagen, type VI, alpha 5 |
14169 |
0.26 |
| chr5_130590704_130590855 | 0.28 |
CDC42SE2 |
CDC42 small effector 2 |
8923 |
0.28 |
| chr18_67623543_67623764 | 0.28 |
CD226 |
CD226 molecule |
253 |
0.95 |
| chr12_13348835_13349296 | 0.27 |
EMP1 |
epithelial membrane protein 1 |
585 |
0.82 |
| chr9_74429580_74429731 | 0.27 |
TMEM2 |
transmembrane protein 2 |
1951 |
0.44 |
| chr2_216650848_216650999 | 0.27 |
ENSG00000212055 |
. |
92719 |
0.09 |
| chr5_35854839_35855004 | 0.27 |
IL7R |
interleukin 7 receptor |
1476 |
0.43 |
| chr12_94805998_94806149 | 0.27 |
CCDC41 |
coiled-coil domain containing 41 |
47649 |
0.14 |
| chr3_24640431_24640582 | 0.27 |
ENSG00000265028 |
. |
77580 |
0.1 |
| chr2_144006550_144006740 | 0.27 |
RP11-190J23.1 |
|
76904 |
0.11 |
| chrX_77039434_77039643 | 0.27 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
2164 |
0.37 |
| chr5_66833336_66833975 | 0.27 |
ENSG00000222939 |
. |
73219 |
0.13 |
| chr11_88087230_88087381 | 0.27 |
CTSC |
cathepsin C |
16350 |
0.28 |
| chr3_156535655_156535806 | 0.27 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
8359 |
0.29 |
| chr3_63954713_63955178 | 0.27 |
ATXN7 |
ataxin 7 |
1525 |
0.34 |
| chr4_109404487_109404709 | 0.27 |
ENSG00000266046 |
. |
93698 |
0.08 |
| chr9_3477118_3477287 | 0.27 |
AL365202.1 |
Uncharacterized protein |
8021 |
0.25 |
| chr13_50650646_50650797 | 0.27 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
5586 |
0.18 |
| chr14_98442707_98443118 | 0.27 |
C14orf64 |
chromosome 14 open reading frame 64 |
1471 |
0.59 |
| chr5_39201497_39202174 | 0.27 |
FYB |
FYN binding protein |
1294 |
0.59 |
| chr3_37214088_37214239 | 0.27 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
1892 |
0.27 |
| chr10_6625751_6625936 | 0.27 |
PRKCQ |
protein kinase C, theta |
3580 |
0.38 |
| chr1_207483654_207483805 | 0.26 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
11124 |
0.27 |
| chr5_72570204_72570355 | 0.26 |
TMEM174 |
transmembrane protein 174 |
101257 |
0.06 |
| chr9_112674232_112674383 | 0.26 |
PALM2 |
paralemmin 2 |
44996 |
0.18 |
| chr2_38826937_38827218 | 0.26 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
91 |
0.96 |
| chr2_160291322_160291473 | 0.26 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
1936 |
0.41 |
| chr14_52688265_52688453 | 0.26 |
PTGDR |
prostaglandin D2 receptor (DP) |
46072 |
0.17 |
| chr15_38852890_38853244 | 0.26 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
587 |
0.75 |
| chr14_65086017_65086168 | 0.26 |
RP11-973N13.3 |
|
22838 |
0.16 |
| chr19_42057151_42057302 | 0.26 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
1340 |
0.4 |
| chr1_161903850_161904001 | 0.26 |
OLFML2B |
olfactomedin-like 2B |
51097 |
0.14 |
| chr14_55541755_55541906 | 0.26 |
MAPK1IP1L |
mitogen-activated protein kinase 1 interacting protein 1-like |
23481 |
0.17 |
| chr7_47333703_47333854 | 0.26 |
TNS3 |
tensin 3 |
13862 |
0.31 |
| chr2_234934004_234934210 | 0.26 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
25216 |
0.2 |
| chr3_99928980_99929378 | 0.26 |
ENSG00000238377 |
. |
11257 |
0.16 |
| chr10_95172691_95172842 | 0.26 |
MYOF |
myoferlin |
69185 |
0.1 |
| chr2_204733780_204733985 | 0.26 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
1073 |
0.62 |
| chr14_106327816_106327967 | 0.26 |
IGHJ6 |
immunoglobulin heavy joining 6 |
1577 |
0.06 |
| chr4_125862018_125862169 | 0.26 |
ANKRD50 |
ankyrin repeat domain 50 |
228206 |
0.02 |
| chr4_148774792_148774943 | 0.26 |
ARHGAP10 |
Rho GTPase activating protein 10 |
28151 |
0.19 |
| chr1_81943750_81943901 | 0.26 |
RP5-837I24.6 |
|
26959 |
0.24 |
| chr21_32567568_32567719 | 0.26 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
65104 |
0.13 |
| chr15_66681568_66681719 | 0.26 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
2488 |
0.18 |
| chr9_130740055_130740787 | 0.26 |
FAM102A |
family with sequence similarity 102, member A |
2371 |
0.18 |
| chr7_86847208_86847359 | 0.26 |
ENSG00000200397 |
. |
745 |
0.59 |
| chr9_802061_802237 | 0.26 |
DMRT1 |
doublesex and mab-3 related transcription factor 1 |
39541 |
0.18 |
| chr17_53314641_53314792 | 0.26 |
HLF |
hepatic leukemia factor |
27657 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.2 | 0.8 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.2 | 0.4 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.2 | 0.5 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.2 | 0.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.6 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.5 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.1 | 0.5 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.1 | 0.4 | GO:0002669 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.3 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.3 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.4 | GO:0002855 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 1.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.2 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.1 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 2.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0032816 | positive regulation of natural killer cell activation(GO:0032816) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.0 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:1904748 | regulation of apoptotic process involved in morphogenesis(GO:1902337) regulation of apoptotic process involved in development(GO:1904748) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.1 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.2 | GO:0060421 | positive regulation of heart growth(GO:0060421) |
| 0.0 | 0.1 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.0 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0045606 | positive regulation of epidermal cell differentiation(GO:0045606) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0006949 | syncytium formation(GO:0006949) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0006273 | lagging strand elongation(GO:0006273) DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.5 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 1.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |