Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
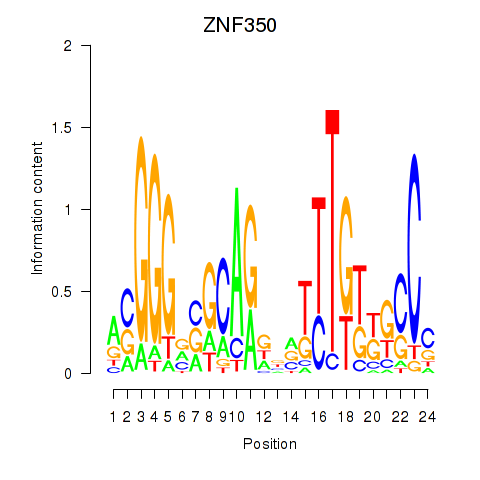
Results for ZNF350
Z-value: 0.80
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | zinc finger protein 350 |
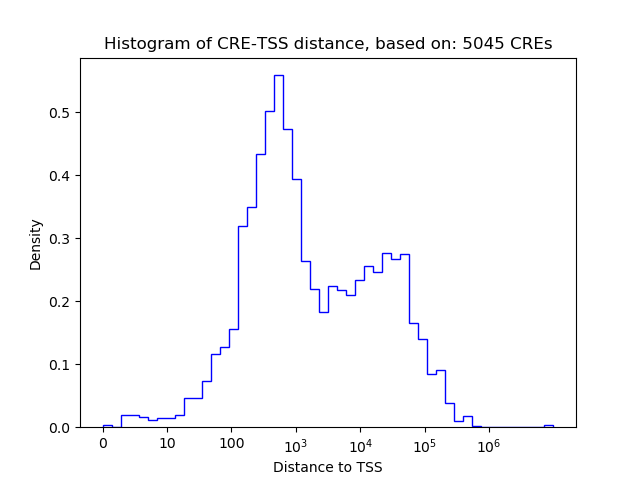
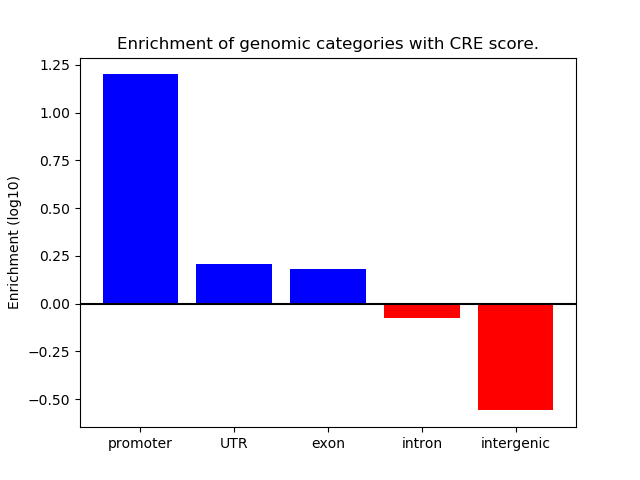
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_52490149_52490417 | ZNF350 | 174 | 0.918947 | -0.57 | 1.1e-01 | Click! |
| chr19_52476505_52476656 | ZNF350 | 1526 | 0.278831 | 0.43 | 2.4e-01 | Click! |
| chr19_52472408_52472559 | ZNF350 | 5623 | 0.130234 | 0.35 | 3.6e-01 | Click! |
| chr19_52468615_52468766 | ZNF350 | 9416 | 0.118112 | 0.34 | 3.8e-01 | Click! |
| chr19_52497579_52497730 | ZNF350 | 7545 | 0.120550 | 0.31 | 4.1e-01 | Click! |
Activity of the ZNF350 motif across conditions
Conditions sorted by the z-value of the ZNF350 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
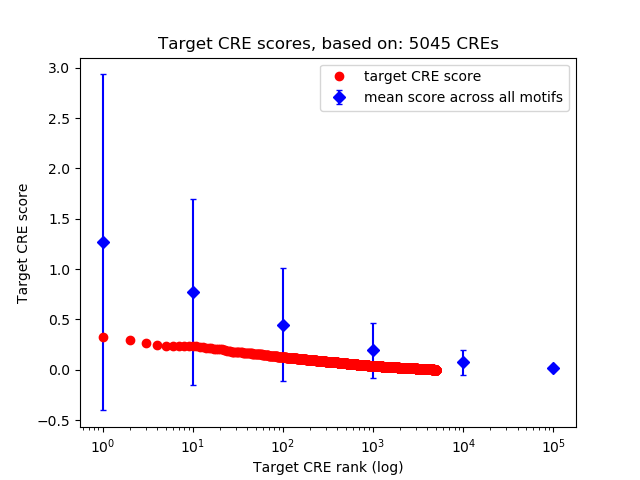
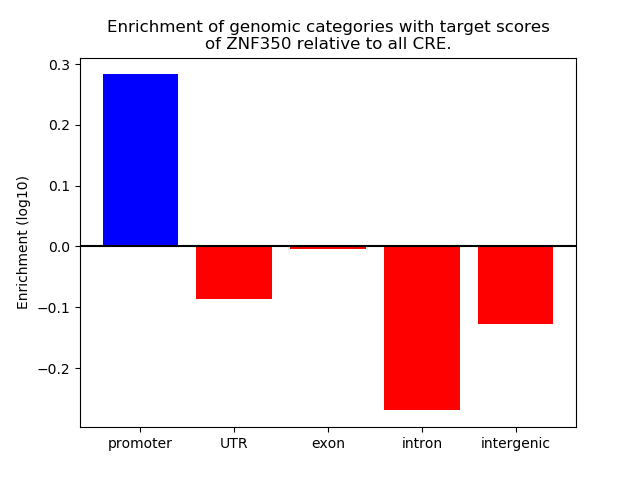
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_129323468_129324020 | 0.33 |
PLXND1 |
plexin D1 |
1917 |
0.3 |
| chr14_106375242_106375393 | 0.30 |
ENSG00000237197 |
. |
465 |
0.3 |
| chr2_206546644_206547002 | 0.27 |
NRP2 |
neuropilin 2 |
401 |
0.91 |
| chr6_56707311_56707814 | 0.25 |
DST |
dystonin |
381 |
0.81 |
| chr6_27759279_27759446 | 0.24 |
HIST1H2BL |
histone cluster 1, H2bl |
16347 |
0.08 |
| chr2_110372829_110373268 | 0.24 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
1137 |
0.38 |
| chr19_49866237_49866676 | 0.24 |
DKKL1 |
dickkopf-like 1 |
125 |
0.85 |
| chr12_41086868_41087222 | 0.24 |
CNTN1 |
contactin 1 |
691 |
0.81 |
| chr10_112116910_112117211 | 0.24 |
SMNDC1 |
survival motor neuron domain containing 1 |
52351 |
0.13 |
| chr17_39940794_39940945 | 0.24 |
JUP |
junction plakoglobin |
538 |
0.62 |
| chr5_132166171_132166684 | 0.23 |
SHROOM1 |
shroom family member 1 |
76 |
0.95 |
| chr10_123873306_123873579 | 0.23 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
888 |
0.71 |
| chr6_56707814_56708209 | 0.22 |
DST |
dystonin |
2 |
0.95 |
| chr20_30457693_30458808 | 0.22 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr21_32931442_32932281 | 0.21 |
AP000251.3 |
|
303 |
0.71 |
| chr13_103052448_103053000 | 0.21 |
FGF14 |
fibroblast growth factor 14 |
1400 |
0.51 |
| chr8_101522130_101522303 | 0.21 |
KB-1615E4.3 |
|
17460 |
0.16 |
| chr17_7119375_7119874 | 0.21 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
820 |
0.27 |
| chr11_75272983_75274329 | 0.21 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
216 |
0.91 |
| chr8_103741653_103741928 | 0.20 |
ENSG00000266799 |
. |
4157 |
0.28 |
| chr20_55204347_55205219 | 0.20 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr7_2728372_2728661 | 0.20 |
AMZ1 |
archaelysin family metallopeptidase 1 |
680 |
0.72 |
| chr2_101435988_101436190 | 0.19 |
NPAS2 |
neuronal PAS domain protein 2 |
525 |
0.8 |
| chr13_113764635_113765129 | 0.19 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
4761 |
0.13 |
| chr8_37552103_37552726 | 0.19 |
ZNF703 |
zinc finger protein 703 |
855 |
0.5 |
| chr7_100464814_100466140 | 0.18 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr21_46875019_46875466 | 0.18 |
COL18A1 |
collagen, type XVIII, alpha 1 |
161 |
0.95 |
| chr2_151342879_151343387 | 0.18 |
RND3 |
Rho family GTPase 3 |
1048 |
0.71 |
| chr6_88875424_88876640 | 0.18 |
CNR1 |
cannabinoid receptor 1 (brain) |
46 |
0.99 |
| chr1_86043549_86043775 | 0.18 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
271 |
0.9 |
| chr2_238395971_238396408 | 0.18 |
MLPH |
melanophilin |
270 |
0.92 |
| chr4_86700818_86700973 | 0.18 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1036 |
0.65 |
| chr4_169552961_169553147 | 0.18 |
PALLD |
palladin, cytoskeletal associated protein |
286 |
0.92 |
| chr9_136858110_136858615 | 0.17 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
636 |
0.73 |
| chr22_29702254_29703430 | 0.17 |
GAS2L1 |
growth arrest-specific 2 like 1 |
171 |
0.91 |
| chr4_3621829_3621980 | 0.17 |
LINC00955 |
long intergenic non-protein coding RNA 955 |
43308 |
0.16 |
| chr5_180631418_180632621 | 0.17 |
TRIM7 |
tripartite motif containing 7 |
153 |
0.85 |
| chr7_3082844_3083007 | 0.17 |
CARD11 |
caspase recruitment domain family, member 11 |
554 |
0.82 |
| chr4_77356334_77356485 | 0.17 |
SHROOM3 |
shroom family member 3 |
156 |
0.95 |
| chr1_109372974_109373275 | 0.17 |
AKNAD1 |
AKNA domain containing 1 |
22215 |
0.15 |
| chr11_62312562_62314176 | 0.17 |
RP11-864I4.4 |
|
102 |
0.64 |
| chr11_27722222_27723091 | 0.16 |
BDNF |
brain-derived neurotrophic factor |
56 |
0.98 |
| chr17_20491916_20492779 | 0.16 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr3_126702505_126703684 | 0.16 |
PLXNA1 |
plexin A1 |
4343 |
0.34 |
| chr15_74428208_74428697 | 0.16 |
ISLR2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
4209 |
0.15 |
| chr4_186065143_186065302 | 0.16 |
SLC25A4 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
827 |
0.63 |
| chr3_118753063_118753796 | 0.16 |
IGSF11 |
immunoglobulin superfamily, member 11 |
146 |
0.97 |
| chr1_98510769_98511355 | 0.16 |
ENSG00000225206 |
. |
155 |
0.98 |
| chr4_114900343_114900754 | 0.16 |
ARSJ |
arylsulfatase family, member J |
309 |
0.93 |
| chr11_8103103_8103531 | 0.16 |
TUB |
tubby bipartite transcription factor |
408 |
0.82 |
| chr9_131903246_131903428 | 0.16 |
PPP2R4 |
protein phosphatase 2A activator, regulatory subunit 4 |
588 |
0.63 |
| chr15_73345145_73345907 | 0.15 |
NEO1 |
neogenin 1 |
575 |
0.85 |
| chr12_113859774_113860410 | 0.15 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr5_1594076_1595069 | 0.15 |
SDHAP3 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3 |
1168 |
0.53 |
| chr9_126807806_126807963 | 0.15 |
RP11-85O21.5 |
|
13081 |
0.17 |
| chr4_75858398_75858601 | 0.15 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
173 |
0.97 |
| chr11_414426_414987 | 0.15 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
242 |
0.84 |
| chr8_8749576_8750657 | 0.15 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
1039 |
0.53 |
| chr2_241458991_241459173 | 0.15 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
862 |
0.54 |
| chr6_105585236_105585620 | 0.15 |
BVES-AS1 |
BVES antisense RNA 1 |
134 |
0.78 |
| chrX_69674453_69675874 | 0.15 |
DLG3 |
discs, large homolog 3 (Drosophila) |
217 |
0.75 |
| chr5_132112834_132113698 | 0.15 |
SEPT8 |
septin 8 |
153 |
0.93 |
| chr22_45898941_45899504 | 0.15 |
FBLN1 |
fibulin 1 |
296 |
0.92 |
| chr1_1283171_1284080 | 0.15 |
DVL1 |
dishevelled segment polarity protein 1 |
867 |
0.32 |
| chr1_16085392_16086186 | 0.15 |
FBLIM1 |
filamin binding LIM protein 1 |
497 |
0.68 |
| chr2_238322230_238322381 | 0.14 |
COL6A3 |
collagen, type VI, alpha 3 |
486 |
0.82 |
| chr15_66999211_66999494 | 0.14 |
SMAD6 |
SMAD family member 6 |
1578 |
0.48 |
| chr7_27223025_27223257 | 0.14 |
HOXA11-AS |
HOXA11 antisense RNA |
996 |
0.23 |
| chr8_22409217_22409843 | 0.14 |
SORBS3 |
sorbin and SH3 domain containing 3 |
322 |
0.81 |
| chr19_39522130_39522840 | 0.14 |
FBXO27 |
F-box protein 27 |
680 |
0.5 |
| chr2_69868222_69868500 | 0.14 |
AAK1 |
AP2 associated kinase 1 |
2425 |
0.31 |
| chr7_37960238_37961117 | 0.14 |
EPDR1 |
ependymin related 1 |
245 |
0.94 |
| chr12_54411200_54411679 | 0.14 |
HOXC4 |
homeobox C4 |
724 |
0.35 |
| chr5_141931000_141931262 | 0.14 |
ENSG00000252831 |
. |
17221 |
0.21 |
| chr21_45663502_45663653 | 0.14 |
ICOSLG |
inducible T-cell co-stimulator ligand |
2728 |
0.16 |
| chr5_38445912_38446355 | 0.14 |
EGFLAM |
EGF-like, fibronectin type III and laminin G domains |
435 |
0.81 |
| chr1_234676868_234677019 | 0.14 |
ENSG00000212144 |
. |
52078 |
0.12 |
| chr2_177502027_177502943 | 0.14 |
ENSG00000252027 |
. |
26919 |
0.25 |
| chr9_109182958_109183109 | 0.14 |
ENSG00000200131 |
. |
259225 |
0.02 |
| chr7_55154680_55154908 | 0.14 |
EGFR |
epidermal growth factor receptor |
22622 |
0.26 |
| chr18_11689181_11689508 | 0.14 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
389 |
0.89 |
| chr8_8242103_8242254 | 0.13 |
SGK223 |
Tyrosine-protein kinase SgK223 |
1830 |
0.46 |
| chr20_57427465_57427903 | 0.13 |
GNAS |
GNAS complex locus |
85 |
0.95 |
| chr2_238117038_238117287 | 0.13 |
AC112715.2 |
Uncharacterized protein |
48572 |
0.16 |
| chr9_98273641_98273827 | 0.13 |
RP11-435O5.4 |
|
2593 |
0.22 |
| chr4_157997002_157997188 | 0.13 |
GLRB |
glycine receptor, beta |
114 |
0.97 |
| chr5_176074525_176075535 | 0.13 |
TSPAN17 |
tetraspanin 17 |
409 |
0.77 |
| chr11_33398103_33398382 | 0.13 |
ENSG00000223134 |
. |
22231 |
0.24 |
| chr9_90691300_90691935 | 0.13 |
ENSG00000271923 |
. |
78367 |
0.1 |
| chr18_35146776_35147412 | 0.13 |
CELF4 |
CUGBP, Elav-like family member 4 |
1094 |
0.67 |
| chr10_102880623_102881444 | 0.13 |
HUG1 |
|
1503 |
0.28 |
| chrY_4868538_4868692 | 0.13 |
PCDH11Y |
protocadherin 11 Y-linked |
348 |
0.92 |
| chr17_76320779_76321257 | 0.13 |
SOCS3 |
suppressor of cytokine signaling 3 |
35137 |
0.12 |
| chr1_240774575_240775223 | 0.13 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
550 |
0.78 |
| chr8_98776052_98776203 | 0.13 |
ENSG00000202399 |
. |
8414 |
0.18 |
| chr1_111148867_111149493 | 0.13 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
205 |
0.94 |
| chr10_48438476_48439044 | 0.13 |
GDF10 |
growth differentiation factor 10 |
216 |
0.93 |
| chr8_41680511_41680662 | 0.13 |
ENSG00000241834 |
. |
17271 |
0.17 |
| chr8_33329381_33329846 | 0.13 |
FUT10 |
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
1024 |
0.5 |
| chr10_73759326_73759477 | 0.13 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
35278 |
0.18 |
| chr2_9239323_9239474 | 0.13 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
95456 |
0.08 |
| chr9_137417944_137418506 | 0.12 |
COL5A1 |
collagen, type V, alpha 1 |
115395 |
0.05 |
| chr11_103407723_103407973 | 0.12 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
178877 |
0.03 |
| chr8_67434316_67434584 | 0.12 |
ENSG00000206949 |
. |
20904 |
0.17 |
| chr2_233388631_233389191 | 0.12 |
CHRND |
cholinergic receptor, nicotinic, delta (muscle) |
1959 |
0.19 |
| chr16_88520261_88521174 | 0.12 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
992 |
0.52 |
| chr2_23608861_23609561 | 0.12 |
KLHL29 |
kelch-like family member 29 |
1123 |
0.67 |
| chr10_16562894_16563258 | 0.12 |
C1QL3 |
complement component 1, q subcomponent-like 3 |
928 |
0.63 |
| chr10_44879737_44880262 | 0.12 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
492 |
0.84 |
| chr8_59904812_59904963 | 0.12 |
RP11-328K2.1 |
|
610 |
0.83 |
| chr6_166721057_166722024 | 0.12 |
PRR18 |
proline rich 18 |
331 |
0.9 |
| chr11_111411049_111411648 | 0.12 |
LAYN |
layilin |
36 |
0.96 |
| chr3_124930827_124931211 | 0.12 |
SLC12A8 |
solute carrier family 12, member 8 |
590 |
0.8 |
| chr2_227662938_227664059 | 0.12 |
IRS1 |
insulin receptor substrate 1 |
977 |
0.58 |
| chr5_141703297_141704624 | 0.12 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
213 |
0.88 |
| chr1_19973947_19974889 | 0.12 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
314 |
0.87 |
| chr2_238537007_238537316 | 0.12 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
942 |
0.55 |
| chr21_27011149_27011750 | 0.12 |
JAM2 |
junctional adhesion molecule 2 |
135 |
0.96 |
| chr2_238321225_238321570 | 0.12 |
COL6A3 |
collagen, type VI, alpha 3 |
1394 |
0.45 |
| chr19_5293472_5293908 | 0.12 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
427 |
0.88 |
| chr12_66122461_66123138 | 0.12 |
HMGA2 |
high mobility group AT-hook 2 |
95112 |
0.08 |
| chr7_36343354_36343505 | 0.12 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
6676 |
0.18 |
| chr1_6452561_6453085 | 0.12 |
ACOT7 |
acyl-CoA thioesterase 7 |
598 |
0.47 |
| chr1_228566293_228567025 | 0.12 |
RP11-245P10.8 |
|
15332 |
0.09 |
| chr10_14215918_14216465 | 0.12 |
RP11-397C18.2 |
|
99908 |
0.08 |
| chr15_39873424_39874712 | 0.12 |
THBS1 |
thrombospondin 1 |
774 |
0.66 |
| chr19_47933466_47933908 | 0.12 |
MEIS3 |
Meis homeobox 3 |
10907 |
0.15 |
| chr6_27661627_27662566 | 0.12 |
ENSG00000238648 |
. |
62908 |
0.08 |
| chr3_111395798_111395949 | 0.12 |
PLCXD2-AS1 |
PLCXD2 antisense RNA 1 |
407 |
0.83 |
| chr6_140303862_140304013 | 0.12 |
ENSG00000252107 |
. |
175894 |
0.03 |
| chr21_39543980_39544194 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
14959 |
0.18 |
| chr20_24449651_24449802 | 0.12 |
SYNDIG1 |
synapse differentiation inducing 1 |
109 |
0.98 |
| chr9_35489468_35490886 | 0.12 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr18_21242120_21242413 | 0.12 |
ANKRD29 |
ankyrin repeat domain 29 |
470 |
0.83 |
| chr11_73028814_73028965 | 0.12 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
6279 |
0.14 |
| chr6_148663623_148664008 | 0.12 |
SASH1 |
SAM and SH3 domain containing 1 |
86 |
0.98 |
| chr12_122461524_122461791 | 0.12 |
BCL7A |
B-cell CLL/lymphoma 7A |
1865 |
0.38 |
| chr15_29862574_29862799 | 0.11 |
FAM189A1 |
family with sequence similarity 189, member A1 |
241 |
0.95 |
| chr17_2118811_2119625 | 0.11 |
AC130689.5 |
|
91 |
0.93 |
| chr2_149634014_149634165 | 0.11 |
KIF5C |
kinesin family member 5C |
1270 |
0.45 |
| chr10_72237920_72238421 | 0.11 |
PALD1 |
phosphatase domain containing, paladin 1 |
407 |
0.85 |
| chr16_72884781_72884932 | 0.11 |
ENSG00000251868 |
. |
28965 |
0.16 |
| chr16_2077067_2078193 | 0.11 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
701 |
0.35 |
| chr2_69240059_69240934 | 0.11 |
ANTXR1 |
anthrax toxin receptor 1 |
33 |
0.98 |
| chr11_118783344_118784212 | 0.11 |
BCL9L |
B-cell CLL/lymphoma 9-like |
2165 |
0.14 |
| chr10_128076891_128077106 | 0.11 |
ADAM12 |
ADAM metallopeptidase domain 12 |
26 |
0.99 |
| chr14_27067583_27067941 | 0.11 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
144 |
0.9 |
| chr12_7036475_7036704 | 0.11 |
ATN1 |
atrophin 1 |
887 |
0.28 |
| chr15_65647670_65647821 | 0.11 |
IGDCC3 |
immunoglobulin superfamily, DCC subclass, member 3 |
504 |
0.77 |
| chr14_30397301_30397598 | 0.11 |
PRKD1 |
protein kinase D1 |
501 |
0.84 |
| chr14_67878511_67879217 | 0.11 |
PLEK2 |
pleckstrin 2 |
53 |
0.97 |
| chr15_30483917_30484165 | 0.11 |
ENSG00000221785 |
. |
46961 |
0.09 |
| chr17_59539414_59539608 | 0.11 |
RP11-15K2.2 |
|
1234 |
0.41 |
| chr5_42424322_42424771 | 0.11 |
GHR |
growth hormone receptor |
520 |
0.88 |
| chr3_179754024_179754473 | 0.11 |
PEX5L |
peroxisomal biogenesis factor 5-like |
310 |
0.94 |
| chr16_85591757_85591908 | 0.11 |
GSE1 |
Gse1 coiled-coil protein |
53183 |
0.13 |
| chr19_17727488_17727789 | 0.11 |
CTD-3149D2.3 |
|
3107 |
0.2 |
| chr19_10542995_10543337 | 0.11 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
55 |
0.95 |
| chr6_86159887_86160764 | 0.11 |
NT5E |
5'-nucleotidase, ecto (CD73) |
498 |
0.87 |
| chr8_145490080_145490245 | 0.11 |
SCXA |
scleraxis homolog A (mouse) |
387 |
0.72 |
| chr1_113392068_113392383 | 0.11 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
86385 |
0.07 |
| chr19_39466589_39467071 | 0.11 |
FBXO17 |
F-box protein 17 |
280 |
0.84 |
| chr15_61520624_61520983 | 0.11 |
RORA |
RAR-related orphan receptor A |
715 |
0.73 |
| chr18_55469401_55470160 | 0.10 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
547 |
0.82 |
| chr12_56137339_56138382 | 0.10 |
GDF11 |
growth differentiation factor 11 |
677 |
0.47 |
| chr11_117185897_117186711 | 0.10 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
210 |
0.87 |
| chr1_99469796_99470269 | 0.10 |
RP5-896L10.1 |
|
200 |
0.74 |
| chr2_66803645_66804000 | 0.10 |
MEIS1 |
Meis homeobox 1 |
67763 |
0.13 |
| chr5_14146856_14147086 | 0.10 |
TRIO |
trio Rho guanine nucleotide exchange factor |
3142 |
0.4 |
| chr13_103450865_103451730 | 0.10 |
KDELC1 |
KDEL (Lys-Asp-Glu-Leu) containing 1 |
60 |
0.54 |
| chr16_70749372_70749620 | 0.10 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
15348 |
0.13 |
| chr5_81685080_81685386 | 0.10 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
84067 |
0.1 |
| chr17_81067040_81067233 | 0.10 |
METRNL |
meteorin, glial cell differentiation regulator-like |
15142 |
0.24 |
| chr21_33784461_33785454 | 0.10 |
EVA1C |
eva-1 homolog C (C. elegans) |
36 |
0.97 |
| chr17_62754772_62754939 | 0.10 |
RP13-104F24.1 |
|
134 |
0.96 |
| chr9_137554285_137554504 | 0.10 |
COL5A1-AS1 |
COL5A1 antisense RNA 1 |
9705 |
0.21 |
| chr1_113243148_113243677 | 0.10 |
RHOC |
ras homolog family member C |
4131 |
0.12 |
| chr7_93931316_93931604 | 0.10 |
COL1A2 |
collagen, type I, alpha 2 |
92413 |
0.09 |
| chr17_71334494_71334713 | 0.10 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
26289 |
0.18 |
| chr2_20795534_20795685 | 0.10 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3301 |
0.28 |
| chr17_42072137_42072373 | 0.10 |
PYY |
peptide YY |
9582 |
0.1 |
| chr6_130690776_130691119 | 0.10 |
TMEM200A |
transmembrane protein 200A |
4068 |
0.25 |
| chr12_132088587_132088738 | 0.10 |
ENSG00000212154 |
. |
56946 |
0.14 |
| chr12_52701821_52702383 | 0.10 |
RP11-845M18.6 |
|
53 |
0.94 |
| chrX_152764727_152764878 | 0.10 |
HAUS7 |
HAUS augmin-like complex, subunit 7 |
3824 |
0.14 |
| chr1_87669967_87670118 | 0.10 |
ENSG00000221222 |
. |
53061 |
0.16 |
| chr7_138666479_138666949 | 0.10 |
KIAA1549 |
KIAA1549 |
650 |
0.77 |
| chr17_58104120_58104612 | 0.10 |
ENSG00000263422 |
. |
15437 |
0.11 |
| chr19_18717102_18717255 | 0.10 |
CRLF1 |
cytokine receptor-like factor 1 |
482 |
0.56 |
| chr17_8230051_8230853 | 0.10 |
ENSG00000212206 |
. |
2449 |
0.13 |
| chr19_7939410_7939591 | 0.10 |
CTD-3193O13.9 |
Protein FLJ22184 |
174 |
0.87 |
| chr7_6296015_6296815 | 0.10 |
CYTH3 |
cytohesin 3 |
15860 |
0.17 |
| chr10_3848106_3848440 | 0.10 |
KLF6 |
Kruppel-like factor 6 |
20800 |
0.22 |
| chr9_115774055_115774548 | 0.10 |
ZFP37 |
ZFP37 zinc finger protein |
44667 |
0.16 |
| chr7_97839825_97840267 | 0.10 |
BHLHA15 |
basic helix-loop-helix family, member a15 |
693 |
0.72 |
| chr6_150921550_150922020 | 0.10 |
RP11-136K14.1 |
|
185 |
0.87 |
| chr6_100676507_100676883 | 0.10 |
RP1-121G13.2 |
|
198299 |
0.03 |
| chr14_106025749_106025900 | 0.10 |
ENSG00000263413 |
. |
30526 |
0.1 |
| chr14_53418107_53419061 | 0.10 |
FERMT2 |
fermitin family member 2 |
569 |
0.83 |
| chr9_130330127_130331202 | 0.10 |
FAM129B |
family with sequence similarity 129, member B |
703 |
0.67 |
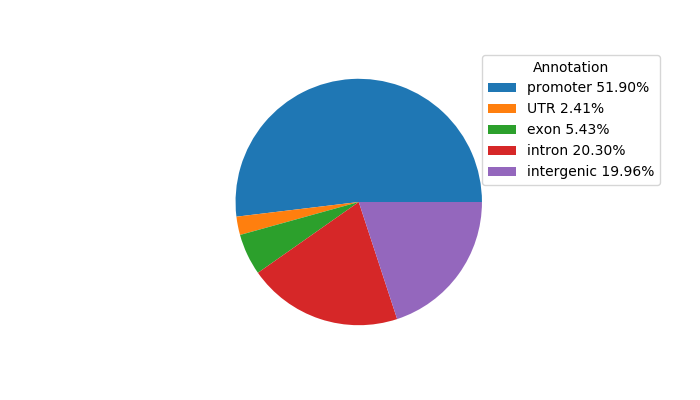
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.2 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0072178 | nephric duct morphogenesis(GO:0072178) mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0033083 | regulation of immature T cell proliferation(GO:0033083) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.0 | GO:0001839 | neural plate morphogenesis(GO:0001839) neural plate development(GO:0001840) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |