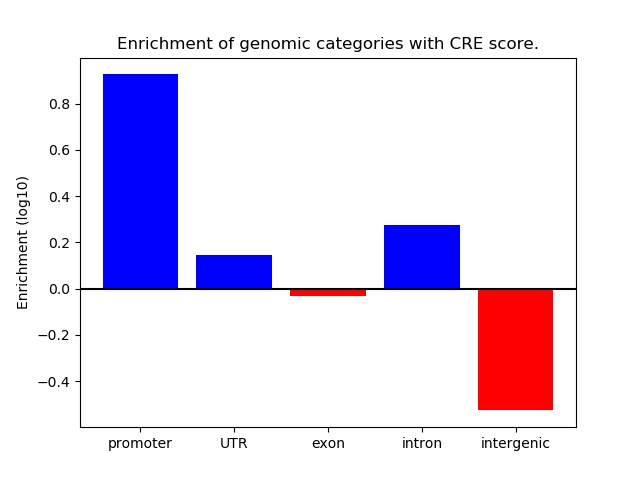
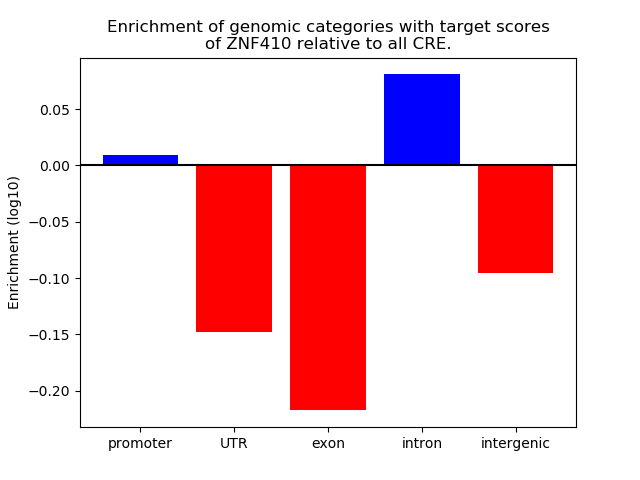
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
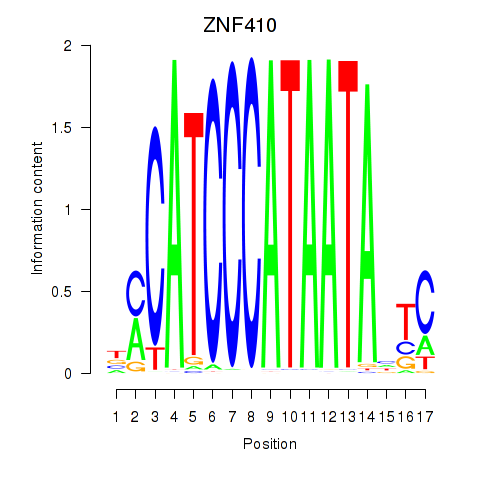
Results for ZNF410
Z-value: 0.53
Transcription factors associated with ZNF410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF410
|
ENSG00000119725.13 | zinc finger protein 410 |
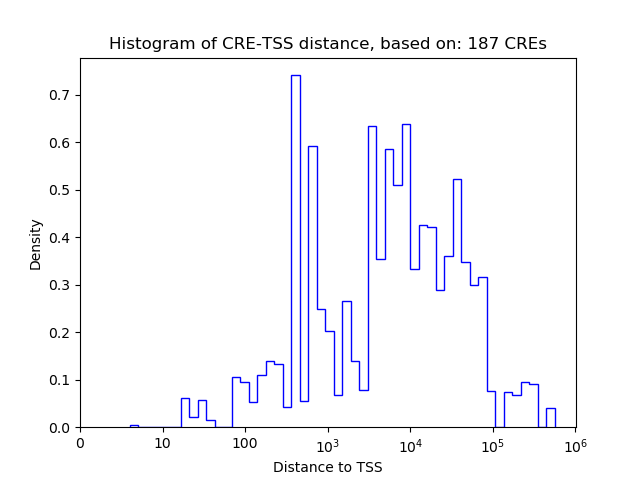
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_74352825_74353319 | ZNF410 | 248 | 0.860110 | -0.26 | 5.0e-01 | Click! |
| chr14_74354228_74354413 | ZNF410 | 559 | 0.618596 | -0.25 | 5.2e-01 | Click! |
| chr14_74353534_74354170 | ZNF410 | 91 | 0.943032 | -0.15 | 7.0e-01 | Click! |
Activity of the ZNF410 motif across conditions
Conditions sorted by the z-value of the ZNF410 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
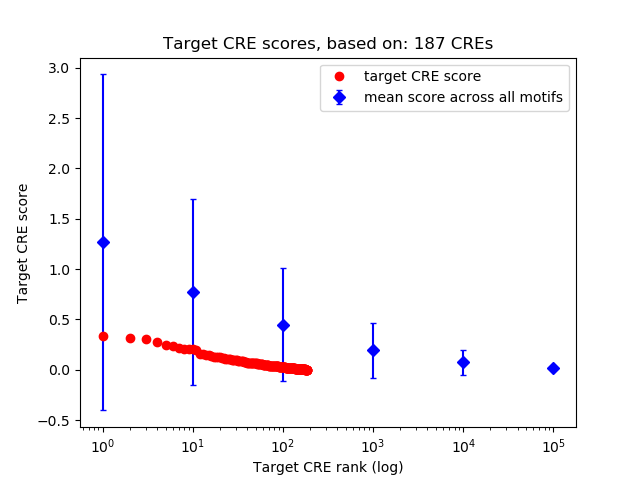
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_21023989_21024197 | 0.34 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7515 |
0.22 |
| chr12_6716856_6717254 | 0.31 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
413 |
0.67 |
| chr8_108507547_108507698 | 0.31 |
ANGPT1 |
angiopoietin 1 |
399 |
0.92 |
| chr10_62331691_62331925 | 0.28 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
613 |
0.85 |
| chr16_50316518_50316669 | 0.24 |
ADCY7 |
adenylate cyclase 7 |
3155 |
0.26 |
| chr2_54825743_54825894 | 0.23 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
40287 |
0.14 |
| chr12_94599811_94599962 | 0.22 |
RP11-74K11.2 |
|
20066 |
0.19 |
| chr7_5682827_5682978 | 0.21 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
37190 |
0.13 |
| chr7_50256147_50256298 | 0.20 |
AC020743.2 |
|
73803 |
0.1 |
| chr12_6722182_6722818 | 0.20 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
5858 |
0.09 |
| chr14_35605928_35606079 | 0.20 |
KIAA0391 |
KIAA0391 |
14038 |
0.19 |
| chr8_53372458_53372609 | 0.16 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
50168 |
0.17 |
| chr12_66274639_66274849 | 0.15 |
RP11-366L20.2 |
Uncharacterized protein |
614 |
0.73 |
| chr11_128586182_128586446 | 0.15 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
20396 |
0.17 |
| chr6_16695295_16695446 | 0.15 |
RP1-151F17.1 |
|
65999 |
0.13 |
| chr6_27104872_27105023 | 0.14 |
HIST1H4I |
histone cluster 1, H4i |
2129 |
0.18 |
| chr3_101555921_101556072 | 0.13 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
9159 |
0.19 |
| chr2_218798521_218798672 | 0.12 |
TNS1 |
tensin 1 |
3289 |
0.29 |
| chr2_201612300_201612519 | 0.12 |
ENSG00000201737 |
. |
27898 |
0.13 |
| chr17_79882093_79882377 | 0.12 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
827 |
0.26 |
| chr1_173175749_173175900 | 0.12 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
628 |
0.81 |
| chr12_69743882_69744071 | 0.12 |
LYZ |
lysozyme |
1810 |
0.3 |
| chr2_201924342_201924493 | 0.11 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
11739 |
0.13 |
| chr3_178866443_178867773 | 0.11 |
PIK3CA |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
207 |
0.94 |
| chr3_101447790_101447941 | 0.11 |
CEP97 |
centrosomal protein 97kDa |
4366 |
0.17 |
| chr19_48747671_48748205 | 0.10 |
CARD8 |
caspase recruitment domain family, member 8 |
3618 |
0.15 |
| chr17_35854577_35854849 | 0.10 |
DUSP14 |
dual specificity phosphatase 14 |
3143 |
0.25 |
| chr9_115981849_115982001 | 0.10 |
FKBP15 |
FK506 binding protein 15, 133kDa |
1673 |
0.28 |
| chr10_114716477_114716628 | 0.10 |
RP11-57H14.2 |
|
4918 |
0.24 |
| chr3_128958405_128958556 | 0.09 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
9969 |
0.15 |
| chr5_36725036_36725307 | 0.09 |
CTD-2353F22.1 |
|
104 |
0.98 |
| chr4_154178413_154178564 | 0.09 |
TRIM2 |
tripartite motif containing 2 |
74 |
0.97 |
| chr4_151509067_151509271 | 0.09 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
6092 |
0.23 |
| chr7_77200176_77200327 | 0.09 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
157 |
0.97 |
| chr2_161243980_161244427 | 0.09 |
ENSG00000252465 |
. |
9276 |
0.21 |
| chr5_111110911_111111062 | 0.08 |
NREP |
neuronal regeneration related protein |
17071 |
0.21 |
| chr6_15421648_15421799 | 0.08 |
JARID2 |
jumonji, AT rich interactive domain 2 |
20634 |
0.24 |
| chr6_170921254_170921775 | 0.08 |
PDCD2 |
programmed cell death 2 |
27766 |
0.19 |
| chr2_236467191_236467342 | 0.07 |
ENSG00000221704 |
. |
13309 |
0.22 |
| chr11_74561071_74561376 | 0.07 |
ENSG00000242999 |
. |
4268 |
0.2 |
| chr10_97501781_97501932 | 0.07 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
13553 |
0.21 |
| chr3_156266030_156266252 | 0.07 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
5773 |
0.24 |
| chr12_46776420_46777113 | 0.07 |
SLC38A2 |
solute carrier family 38, member 2 |
10116 |
0.24 |
| chr5_176901024_176901292 | 0.07 |
DBN1 |
drebrin 1 |
244 |
0.85 |
| chr5_124090344_124090580 | 0.07 |
ZNF608 |
zinc finger protein 608 |
5962 |
0.15 |
| chr14_73927569_73927740 | 0.07 |
ENSG00000251393 |
. |
1475 |
0.32 |
| chr2_48599257_48599408 | 0.07 |
ENSG00000251889 |
. |
31367 |
0.18 |
| chr1_39624513_39624771 | 0.07 |
ENSG00000222378 |
. |
4674 |
0.21 |
| chr4_40561734_40561885 | 0.06 |
RBM47 |
RNA binding motif protein 47 |
43819 |
0.15 |
| chr9_83413109_83413260 | 0.06 |
ENSG00000221581 |
. |
63940 |
0.16 |
| chr13_21654404_21654812 | 0.06 |
LATS2 |
large tumor suppressor kinase 2 |
18922 |
0.14 |
| chr11_57334584_57335055 | 0.06 |
UBE2L6 |
ubiquitin-conjugating enzyme E2L 6 |
20 |
0.96 |
| chr16_17087479_17087630 | 0.06 |
CTD-2576D5.4 |
|
140807 |
0.05 |
| chr10_111659026_111659177 | 0.06 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
8476 |
0.22 |
| chr6_1841402_1841553 | 0.06 |
FOXC1 |
forkhead box C1 |
230796 |
0.02 |
| chr5_14154391_14154790 | 0.06 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10761 |
0.32 |
| chr14_56584316_56584807 | 0.06 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
29 |
0.98 |
| chr13_78410734_78410885 | 0.05 |
EDNRB |
endothelin receptor type B |
82136 |
0.08 |
| chr11_10064267_10064418 | 0.05 |
SBF2 |
SET binding factor 2 |
49691 |
0.16 |
| chr3_119922105_119922256 | 0.05 |
ENSG00000206721 |
. |
6987 |
0.22 |
| chr10_116582241_116583014 | 0.05 |
FAM160B1 |
family with sequence similarity 160, member B1 |
1098 |
0.64 |
| chr2_45794929_45795131 | 0.05 |
SRBD1 |
S1 RNA binding domain 1 |
124 |
0.96 |
| chr6_163826222_163826373 | 0.05 |
QKI |
QKI, KH domain containing, RNA binding |
9378 |
0.32 |
| chr19_1064805_1065015 | 0.05 |
HMHA1 |
histocompatibility (minor) HA-1 |
1012 |
0.32 |
| chr8_127989397_127989548 | 0.05 |
ENSG00000212451 |
. |
305705 |
0.01 |
| chr8_86371428_86371579 | 0.05 |
RP11-317J10.2 |
|
3780 |
0.21 |
| chr3_63957905_63958056 | 0.05 |
ATXN7 |
ataxin 7 |
4560 |
0.17 |
| chr6_85134970_85135121 | 0.05 |
KIAA1009 |
KIAA1009 |
197692 |
0.03 |
| chr14_67828227_67828378 | 0.04 |
EIF2S1 |
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
1029 |
0.38 |
| chr6_41666551_41666729 | 0.04 |
TFEB |
transcription factor EB |
7038 |
0.13 |
| chr10_7670560_7670711 | 0.04 |
ITIH5 |
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
9012 |
0.26 |
| chr12_84274084_84274235 | 0.04 |
ENSG00000221148 |
. |
303068 |
0.01 |
| chr5_40615963_40616141 | 0.04 |
ENSG00000199552 |
. |
39013 |
0.18 |
| chrX_143576959_143577110 | 0.04 |
ENSG00000201912 |
. |
561594 |
0.0 |
| chr11_15179065_15179216 | 0.04 |
INSC |
inscuteable homolog (Drosophila) |
8486 |
0.3 |
| chr2_65291860_65292299 | 0.04 |
CEP68 |
centrosomal protein 68kDa |
8480 |
0.18 |
| chr20_36629792_36630028 | 0.04 |
TTI1 |
TELO2 interacting protein 1 |
12395 |
0.17 |
| chrX_153637082_153637306 | 0.04 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
418 |
0.63 |
| chr5_172261659_172261898 | 0.04 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
364 |
0.87 |
| chr8_123440615_123440780 | 0.04 |
ENSG00000238901 |
. |
242833 |
0.02 |
| chr12_123199549_123199700 | 0.04 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
1815 |
0.27 |
| chr12_109118598_109118801 | 0.04 |
CORO1C |
coronin, actin binding protein, 1C |
5628 |
0.17 |
| chr2_202519598_202519749 | 0.04 |
ENSG00000222972 |
. |
8234 |
0.16 |
| chr5_132385127_132385278 | 0.04 |
HSPA4 |
heat shock 70kDa protein 4 |
2452 |
0.27 |
| chr1_206137311_206137809 | 0.04 |
RP11-312O7.2 |
|
268 |
0.52 |
| chrX_24073135_24073641 | 0.04 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
300 |
0.91 |
| chr4_22516047_22517223 | 0.04 |
GPR125 |
G protein-coupled receptor 125 |
985 |
0.72 |
| chr7_17018287_17018438 | 0.03 |
AGR3 |
anterior gradient 3 |
96751 |
0.08 |
| chr9_116384320_116384471 | 0.03 |
RGS3 |
regulator of G-protein signaling 3 |
28629 |
0.19 |
| chr9_41563760_41563911 | 0.03 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
63156 |
0.15 |
| chr19_36618863_36619073 | 0.03 |
CAPNS1 |
calpain, small subunit 1 |
11509 |
0.09 |
| chr20_23067023_23067351 | 0.03 |
CD93 |
CD93 molecule |
210 |
0.93 |
| chr1_98496294_98496445 | 0.03 |
ENSG00000225206 |
. |
14538 |
0.29 |
| chr22_37679105_37679462 | 0.03 |
CYTH4 |
cytohesin 4 |
755 |
0.62 |
| chr12_13499997_13500148 | 0.03 |
C12orf36 |
chromosome 12 open reading frame 36 |
29292 |
0.21 |
| chr12_65071079_65071230 | 0.03 |
AC025262.1 |
Mesenchymal stem cell protein DSC96; Uncharacterized protein |
19175 |
0.13 |
| chr13_28997424_28997626 | 0.03 |
FLT1 |
fms-related tyrosine kinase 1 |
71707 |
0.12 |
| chr15_85295872_85296023 | 0.03 |
RP11-7M10.2 |
|
406 |
0.79 |
| chr1_225942852_225943140 | 0.03 |
ENSG00000223306 |
. |
14019 |
0.14 |
| chr1_175323766_175323917 | 0.03 |
RP3-518E13.2 |
|
47487 |
0.16 |
| chr1_43416112_43416263 | 0.03 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
8313 |
0.19 |
| chr7_87504669_87504826 | 0.02 |
DBF4 |
DBF4 homolog (S. cerevisiae) |
797 |
0.47 |
| chr5_178956987_178957198 | 0.02 |
RUFY1 |
RUN and FYVE domain containing 1 |
20467 |
0.17 |
| chr13_42151532_42151903 | 0.02 |
ENSG00000264190 |
. |
9186 |
0.26 |
| chr9_39418756_39418907 | 0.02 |
CTD-2173L22.4 |
|
42860 |
0.18 |
| chr10_61661518_61661669 | 0.02 |
CCDC6 |
coiled-coil domain containing 6 |
4821 |
0.3 |
| chrX_69512839_69512990 | 0.02 |
KIF4A |
kinesin family member 4A |
2974 |
0.15 |
| chr5_119488092_119488243 | 0.02 |
ENSG00000251975 |
. |
185181 |
0.03 |
| chr10_127511437_127512239 | 0.02 |
UROS |
uroporphyrinogen III synthase |
21 |
0.79 |
| chr8_74904321_74904899 | 0.02 |
LY96 |
lymphocyte antigen 96 |
949 |
0.5 |
| chr8_94752281_94753315 | 0.02 |
RBM12B |
RNA binding motif protein 12B |
148 |
0.72 |
| chr7_13920022_13920238 | 0.02 |
ETV1 |
ets variant 1 |
105936 |
0.08 |
| chr17_57701437_57701588 | 0.02 |
CLTC |
clathrin, heavy chain (Hc) |
4219 |
0.23 |
| chrX_64925761_64925912 | 0.02 |
MSN |
moesin |
38299 |
0.22 |
| chr10_134363355_134363506 | 0.02 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
11787 |
0.21 |
| chr9_39948000_39948151 | 0.02 |
SPATA31A2 |
SPATA31 subfamily A, member 2 |
63100 |
0.14 |
| chr2_62638898_62639095 | 0.02 |
ENSG00000241625 |
. |
79317 |
0.09 |
| chr14_92571968_92572886 | 0.02 |
ATXN3 |
ataxin 3 |
479 |
0.77 |
| chr17_66437196_66437347 | 0.02 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
7559 |
0.21 |
| chr11_8008996_8010000 | 0.02 |
EIF3F |
eukaryotic translation initiation factor 3, subunit F |
610 |
0.64 |
| chr2_210868078_210868268 | 0.02 |
RPE |
ribulose-5-phosphate-3-epimerase |
787 |
0.63 |
| chr1_16177042_16177194 | 0.02 |
RP11-169K16.9 |
|
2476 |
0.21 |
| chr13_46741782_46741933 | 0.02 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
797 |
0.56 |
| chr15_40226407_40226907 | 0.02 |
EIF2AK4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
280 |
0.88 |
| chr11_85778298_85779603 | 0.02 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
872 |
0.65 |
| chr9_113598301_113598563 | 0.02 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
60805 |
0.12 |
| chr19_13023816_13024331 | 0.02 |
SYCE2 |
synaptonemal complex central element protein 2 |
6017 |
0.06 |
| chr12_114284871_114285022 | 0.02 |
RP11-780K2.1 |
|
85428 |
0.1 |
| chr5_72111935_72112277 | 0.02 |
TNPO1 |
transportin 1 |
33 |
0.8 |
| chr1_40847512_40847991 | 0.02 |
SMAP2 |
small ArfGAP2 |
7431 |
0.18 |
| chr8_134090212_134090363 | 0.01 |
SLA |
Src-like-adaptor |
17684 |
0.22 |
| chr3_125635457_125636442 | 0.01 |
FAM86JP |
family with sequence similarity 86, member J, pseudogene |
482 |
0.8 |
| chr15_102061524_102061675 | 0.01 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
31412 |
0.2 |
| chr9_129252632_129253068 | 0.01 |
ENSG00000221768 |
. |
40420 |
0.14 |
| chr3_168825087_168825238 | 0.01 |
MECOM |
MDS1 and EVI1 complex locus |
20660 |
0.29 |
| chr1_221209086_221209419 | 0.01 |
HLX |
H2.0-like homeobox |
154668 |
0.04 |
| chr1_143913778_143914315 | 0.01 |
FAM72D |
family with sequence similarity 72, member D |
903 |
0.56 |
| chr18_32663220_32663371 | 0.01 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
41681 |
0.2 |
| chr19_9524867_9525018 | 0.01 |
ZNF266 |
zinc finger protein 266 |
4815 |
0.19 |
| chr7_37759942_37760093 | 0.01 |
GPR141 |
G protein-coupled receptor 141 |
19979 |
0.23 |
| chr14_21851512_21852227 | 0.01 |
SUPT16H |
suppressor of Ty 16 homolog (S. cerevisiae) |
271 |
0.69 |
| chr5_95144892_95145187 | 0.01 |
GLRX |
glutaredoxin (thioltransferase) |
13376 |
0.15 |
| chr6_160450858_160451009 | 0.01 |
AIRN |
antisense of IGF2R non-protein coding RNA |
22237 |
0.21 |
| chr8_37888134_37889006 | 0.01 |
EIF4EBP1 |
eukaryotic translation initiation factor 4E binding protein 1 |
711 |
0.62 |
| chr15_52788839_52789035 | 0.01 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
32101 |
0.18 |
| chr22_46134071_46134222 | 0.01 |
ATXN10 |
ataxin 10 |
473 |
0.84 |
| chr6_71544283_71544434 | 0.01 |
SMAP1 |
small ArfGAP 1 |
61266 |
0.15 |
| chr10_70165992_70166978 | 0.01 |
RUFY2 |
RUN and FYVE domain containing 2 |
461 |
0.8 |
| chr9_14309575_14309740 | 0.01 |
NFIB |
nuclear factor I/B |
1645 |
0.46 |
| chr14_71066645_71067347 | 0.01 |
MED6 |
mediator complex subunit 6 |
364 |
0.87 |
| chr13_46742646_46742820 | 0.01 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
79 |
0.95 |
| chr6_121760674_121760877 | 0.01 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
3937 |
0.22 |
| chr14_92874571_92874722 | 0.01 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
77 |
0.98 |
| chr1_231671479_231671630 | 0.01 |
TSNAX |
translin-associated factor X |
7153 |
0.2 |
| chr17_13689312_13689463 | 0.01 |
ENSG00000236088 |
. |
6480 |
0.33 |
| chr14_32842058_32842209 | 0.01 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
43527 |
0.15 |
| chr13_86099472_86099623 | 0.01 |
ENSG00000207012 |
. |
59441 |
0.17 |
| chr19_58521386_58521539 | 0.01 |
ZNF606 |
zinc finger protein 606 |
6745 |
0.11 |
| chr2_60755895_60756046 | 0.01 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
24570 |
0.2 |
| chr4_37978441_37978791 | 0.01 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
315 |
0.92 |
| chr3_88483370_88483521 | 0.01 |
ENSG00000207316 |
. |
3104 |
0.41 |
| chr9_80408593_80408744 | 0.01 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
29247 |
0.25 |
| chr1_27079827_27079978 | 0.00 |
ENSG00000266138 |
. |
10213 |
0.13 |
| chr13_103353320_103353471 | 0.00 |
METTL21C |
methyltransferase like 21C |
6541 |
0.16 |
| chr1_87803835_87803986 | 0.00 |
LMO4 |
LIM domain only 4 |
6559 |
0.32 |
| chr8_93547538_93547689 | 0.00 |
ENSG00000221172 |
. |
99950 |
0.09 |
| chr18_55469401_55470160 | 0.00 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
547 |
0.82 |
| chr16_14800704_14800855 | 0.00 |
RP11-82O18.2 |
|
133 |
0.93 |
| chr12_95596867_95597053 | 0.00 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
14195 |
0.18 |
| chr12_57940971_57941264 | 0.00 |
DCTN2 |
dynactin 2 (p50) |
3 |
0.94 |
| chr3_102392529_102392885 | 0.00 |
ENSG00000201065 |
. |
81307 |
0.12 |
| chr8_42409378_42409529 | 0.00 |
SMIM19 |
small integral membrane protein 19 |
7882 |
0.18 |
| chr14_24611030_24611338 | 0.00 |
EMC9 |
ER membrane protein complex subunit 9 |
387 |
0.61 |
| chrX_78412852_78413003 | 0.00 |
GPR174 |
G protein-coupled receptor 174 |
13542 |
0.31 |
| chr18_43554027_43554236 | 0.00 |
EPG5 |
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
6891 |
0.21 |
| chr16_53751617_53751768 | 0.00 |
FTO |
fat mass and obesity associated |
13598 |
0.22 |
| chr2_71763544_71763695 | 0.00 |
DYSF |
dysferlin |
69787 |
0.12 |
| chr12_121726859_121727365 | 0.00 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
7377 |
0.21 |
| chr12_100988036_100988187 | 0.00 |
GAS2L3 |
growth arrest-specific 2 like 3 |
715 |
0.75 |
| chr12_62657622_62657800 | 0.00 |
USP15 |
ubiquitin specific peptidase 15 |
3502 |
0.25 |
| chr6_107816329_107816751 | 0.00 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
5378 |
0.23 |
| chr10_5509058_5509209 | 0.00 |
NET1 |
neuroepithelial cell transforming 1 |
20559 |
0.14 |
| chr11_114034692_114034843 | 0.00 |
ENSG00000221112 |
. |
77116 |
0.09 |
| chr10_126368779_126368930 | 0.00 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
23340 |
0.19 |
| chr17_37557069_37557922 | 0.00 |
FBXL20 |
F-box and leucine-rich repeat protein 20 |
381 |
0.59 |
| chr2_179279129_179279546 | 0.00 |
AC009948.5 |
|
587 |
0.72 |
| chr15_48469342_48470564 | 0.00 |
MYEF2 |
myelin expression factor 2 |
597 |
0.66 |
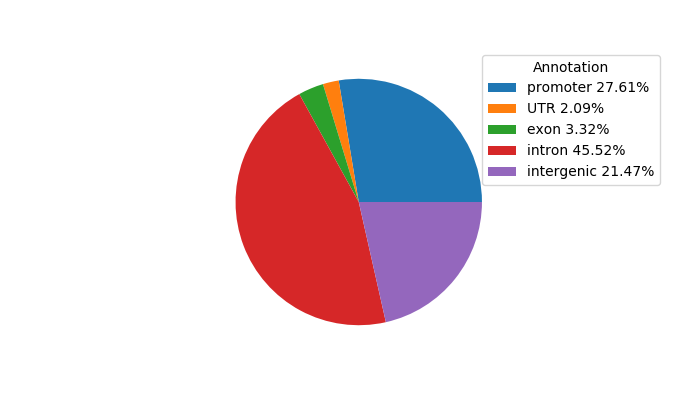
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |