Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
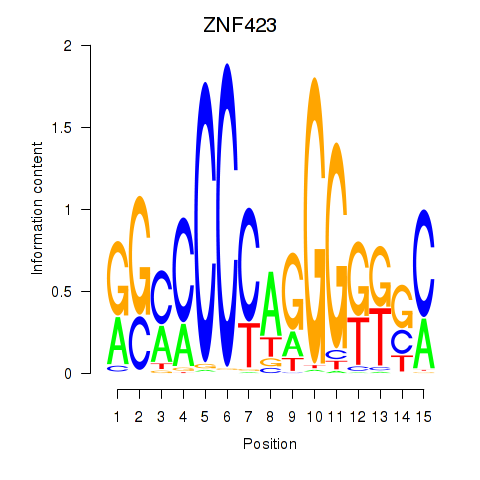
Results for ZNF423
Z-value: 0.63
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | zinc finger protein 423 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_49629926_49630278 | ZNF423 | 44661 | 0.181152 | 0.85 | 4.0e-03 | Click! |
| chr16_49654580_49654731 | ZNF423 | 20108 | 0.244998 | 0.75 | 1.9e-02 | Click! |
| chr16_49697543_49697694 | ZNF423 | 518 | 0.835648 | 0.72 | 2.8e-02 | Click! |
| chr16_49862024_49862175 | ZNF423 | 1178 | 0.547025 | 0.71 | 3.0e-02 | Click! |
| chr16_49698067_49698218 | ZNF423 | 6 | 0.984072 | 0.69 | 4.1e-02 | Click! |
Activity of the ZNF423 motif across conditions
Conditions sorted by the z-value of the ZNF423 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
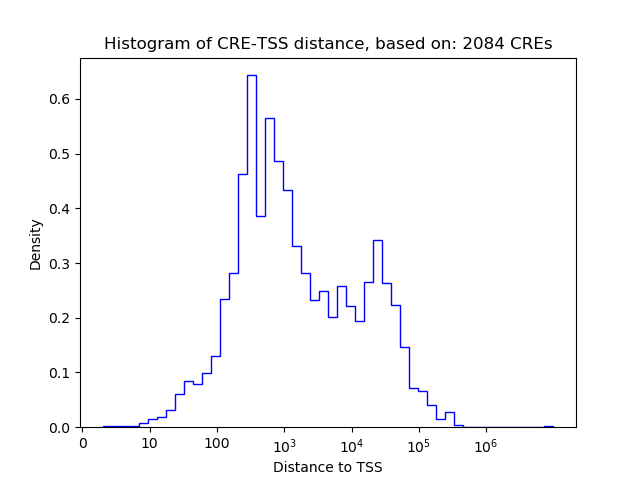
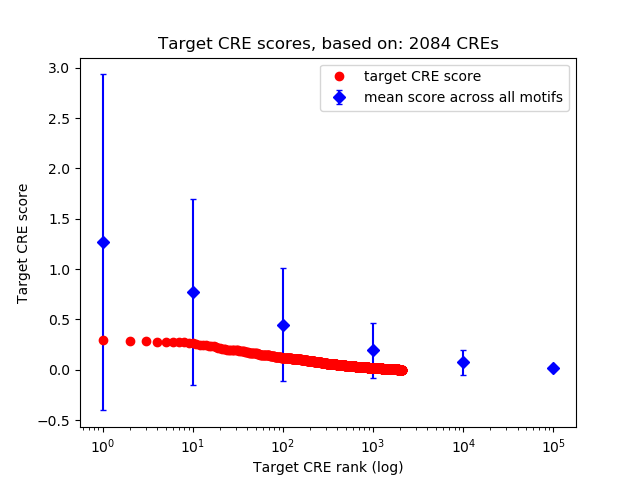
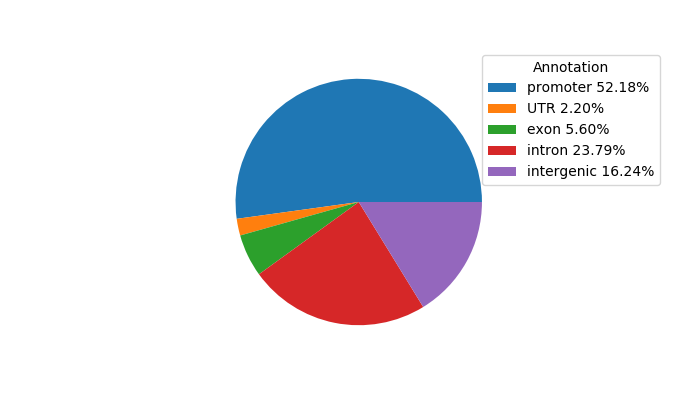
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_150464397_150464642 | 0.30 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
307 |
0.91 |
| chr3_114173030_114173441 | 0.29 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
295 |
0.94 |
| chr13_95619763_95620412 | 0.28 |
ENSG00000252335 |
. |
51402 |
0.18 |
| chr22_30641304_30641455 | 0.28 |
RP1-102K2.8 |
|
605 |
0.52 |
| chr1_14924992_14926197 | 0.28 |
KAZN |
kazrin, periplakin interacting protein |
381 |
0.93 |
| chr17_17725795_17725961 | 0.28 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
1054 |
0.42 |
| chr12_66216582_66216994 | 0.27 |
HMGA2 |
high mobility group AT-hook 2 |
1123 |
0.55 |
| chr1_20915430_20915891 | 0.27 |
CDA |
cytidine deaminase |
219 |
0.93 |
| chr10_63212172_63213035 | 0.27 |
RP11-809M12.1 |
|
206 |
0.83 |
| chr19_41725215_41725482 | 0.26 |
AXL |
AXL receptor tyrosine kinase |
208 |
0.89 |
| chr6_150464730_150464955 | 0.25 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
630 |
0.74 |
| chrX_39548256_39548819 | 0.25 |
ENSG00000263730 |
. |
28067 |
0.24 |
| chr3_133614813_133615159 | 0.24 |
RAB6B |
RAB6B, member RAS oncogene family |
306 |
0.92 |
| chr13_114541163_114541465 | 0.24 |
GAS6 |
growth arrest-specific 6 |
2297 |
0.34 |
| chr1_27928484_27928752 | 0.24 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
1484 |
0.34 |
| chr8_89340038_89340774 | 0.24 |
MMP16 |
matrix metallopeptidase 16 (membrane-inserted) |
152 |
0.73 |
| chr20_6032553_6033205 | 0.23 |
LRRN4 |
leucine rich repeat neuronal 4 |
1816 |
0.35 |
| chr2_241860515_241860848 | 0.22 |
AC104809.3 |
Protein LOC728763 |
1305 |
0.38 |
| chr19_45147190_45148055 | 0.22 |
PVR |
poliovirus receptor |
225 |
0.9 |
| chr2_127976513_127977351 | 0.21 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
722 |
0.74 |
| chr2_113593127_113593420 | 0.21 |
IL1B |
interleukin 1, beta |
738 |
0.63 |
| chr19_1240029_1240222 | 0.21 |
ATP5D |
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
1624 |
0.18 |
| chr19_36024645_36024970 | 0.20 |
GAPDHS |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
493 |
0.62 |
| chr1_208132304_208132634 | 0.20 |
CD34 |
CD34 molecule |
47722 |
0.19 |
| chr11_64122826_64123177 | 0.20 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
3619 |
0.11 |
| chr20_57090585_57090835 | 0.20 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
523 |
0.83 |
| chr19_54483356_54483962 | 0.20 |
ENSG00000215998 |
. |
1902 |
0.19 |
| chr7_20370368_20370519 | 0.19 |
ITGB8 |
integrin, beta 8 |
118 |
0.93 |
| chr4_74735225_74735723 | 0.19 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
353 |
0.85 |
| chr1_65991788_65991976 | 0.19 |
LEPR |
leptin receptor |
39347 |
0.14 |
| chr10_72976782_72977411 | 0.19 |
UNC5B-AS1 |
UNC5B antisense RNA 1 |
486 |
0.73 |
| chr22_29598883_29599280 | 0.19 |
EMID1 |
EMI domain containing 1 |
2759 |
0.2 |
| chr4_156681206_156681472 | 0.19 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
728 |
0.72 |
| chr7_23508285_23508467 | 0.19 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
1710 |
0.33 |
| chr2_218874347_218874698 | 0.19 |
TNS1 |
tensin 1 |
6804 |
0.19 |
| chr22_46482353_46482522 | 0.19 |
FLJ27365 |
hsa-mir-4763 |
554 |
0.6 |
| chr13_96296247_96296898 | 0.18 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
372 |
0.89 |
| chr16_88870757_88870966 | 0.18 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
1240 |
0.28 |
| chr12_81330378_81331020 | 0.17 |
LIN7A |
lin-7 homolog A (C. elegans) |
784 |
0.37 |
| chr17_8295251_8295402 | 0.17 |
RNF222 |
ring finger protein 222 |
1481 |
0.24 |
| chr21_35831656_35831815 | 0.17 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
167 |
0.94 |
| chr7_43288672_43288925 | 0.17 |
AC004692.4 |
|
43 |
0.98 |
| chr7_73704780_73705014 | 0.17 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
1092 |
0.51 |
| chr2_97136526_97136687 | 0.17 |
NEURL3 |
neuralized E3 ubiquitin protein ligase 3 |
30055 |
0.16 |
| chr19_44905150_44906080 | 0.17 |
ZNF285 |
zinc finger protein 285 |
126 |
0.54 |
| chr8_55065813_55066181 | 0.17 |
ENSG00000251835 |
. |
555 |
0.79 |
| chr4_74809740_74810515 | 0.17 |
PF4 |
platelet factor 4 |
37714 |
0.11 |
| chr17_71301523_71301822 | 0.17 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
5875 |
0.2 |
| chr16_4424352_4424847 | 0.16 |
VASN |
vasorin |
2750 |
0.17 |
| chr19_10947512_10947663 | 0.16 |
C19orf38 |
chromosome 19 open reading frame 38 |
336 |
0.63 |
| chr4_62067084_62067588 | 0.16 |
LPHN3 |
latrophilin 3 |
360 |
0.94 |
| chr4_74903896_74904220 | 0.16 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
348 |
0.83 |
| chr7_47620495_47620884 | 0.16 |
TNS3 |
tensin 3 |
540 |
0.86 |
| chr15_101629242_101629566 | 0.16 |
RP11-505E24.2 |
|
3133 |
0.3 |
| chr7_37960238_37961117 | 0.15 |
EPDR1 |
ependymin related 1 |
245 |
0.94 |
| chr10_106079408_106079559 | 0.15 |
RP11-127L20.5 |
|
7584 |
0.13 |
| chr10_114712061_114712425 | 0.15 |
RP11-57H14.2 |
|
609 |
0.69 |
| chr17_37729930_37730438 | 0.15 |
ENSG00000222777 |
. |
27687 |
0.12 |
| chr4_74734701_74735215 | 0.15 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
163 |
0.94 |
| chr13_33804184_33804599 | 0.15 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
24248 |
0.2 |
| chr5_9544908_9545592 | 0.15 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
937 |
0.56 |
| chr19_17579873_17580031 | 0.15 |
SLC27A1 |
solute carrier family 27 (fatty acid transporter), member 1 |
374 |
0.71 |
| chr5_167792475_167792729 | 0.15 |
WWC1 |
WW and C2 domain containing 1 |
5824 |
0.29 |
| chrX_19140154_19140583 | 0.15 |
GPR64 |
G protein-coupled receptor 64 |
211 |
0.97 |
| chr18_500844_501279 | 0.15 |
COLEC12 |
collectin sub-family member 12 |
339 |
0.89 |
| chr16_70719186_70719575 | 0.15 |
MTSS1L |
metastasis suppressor 1-like |
589 |
0.67 |
| chr20_1298176_1298327 | 0.15 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
3934 |
0.16 |
| chr1_78354680_78354975 | 0.14 |
NEXN-AS1 |
NEXN antisense RNA 1 |
397 |
0.54 |
| chr11_66082073_66082224 | 0.14 |
RP11-867G23.13 |
|
1824 |
0.12 |
| chr18_11149170_11149978 | 0.14 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
987 |
0.72 |
| chr9_140301738_140301959 | 0.14 |
ENSG00000272272 |
. |
13748 |
0.11 |
| chr17_7483283_7483711 | 0.14 |
CD68 |
CD68 molecule |
316 |
0.62 |
| chr13_114541513_114541687 | 0.14 |
GAS6 |
growth arrest-specific 6 |
2583 |
0.32 |
| chr1_54844833_54844984 | 0.14 |
SSBP3 |
single stranded DNA binding protein 3 |
26269 |
0.19 |
| chr9_18438384_18438699 | 0.14 |
ADAMTSL1 |
ADAMTS-like 1 |
35351 |
0.23 |
| chr9_112542645_112542955 | 0.13 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
31 |
0.37 |
| chr7_100880679_100880896 | 0.13 |
CLDN15 |
claudin 15 |
254 |
0.85 |
| chr17_58164259_58164533 | 0.13 |
HEATR6 |
HEAT repeat containing 6 |
8104 |
0.14 |
| chr22_50698815_50699396 | 0.13 |
MAPK12 |
mitogen-activated protein kinase 12 |
632 |
0.49 |
| chr13_27757022_27757206 | 0.13 |
USP12-AS2 |
USP12 antisense RNA 2 (head to head) |
10718 |
0.14 |
| chr5_115911326_115911646 | 0.13 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
856 |
0.75 |
| chr12_43945597_43946279 | 0.13 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
214 |
0.97 |
| chr1_156072742_156072967 | 0.13 |
LMNA |
lamin A/C |
11607 |
0.1 |
| chr11_69460044_69460271 | 0.13 |
CCND1 |
cyclin D1 |
4183 |
0.25 |
| chr5_176935395_176936034 | 0.13 |
DOK3 |
docking protein 3 |
1113 |
0.32 |
| chr2_14772882_14773213 | 0.13 |
FAM84A |
family with sequence similarity 84, member A |
223 |
0.94 |
| chr7_101268427_101268664 | 0.13 |
MYL10 |
myosin, light chain 10, regulatory |
4031 |
0.33 |
| chr12_32551927_32552214 | 0.13 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
393 |
0.86 |
| chr7_136555280_136555665 | 0.13 |
CHRM2 |
cholinergic receptor, muscarinic 2 |
1603 |
0.4 |
| chr5_82768204_82768925 | 0.13 |
VCAN |
versican |
820 |
0.75 |
| chr6_139014672_139015008 | 0.13 |
RP11-390P2.4 |
|
616 |
0.63 |
| chr7_101460813_101461304 | 0.13 |
CUX1 |
cut-like homeobox 1 |
138 |
0.97 |
| chr2_26785075_26785548 | 0.12 |
C2orf70 |
chromosome 2 open reading frame 70 |
139 |
0.96 |
| chr2_28601499_28601815 | 0.12 |
FOSL2 |
FOS-like antigen 2 |
14012 |
0.17 |
| chr15_31631055_31631206 | 0.12 |
KLF13 |
Kruppel-like factor 13 |
1006 |
0.69 |
| chr1_12656148_12656570 | 0.12 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
38 |
0.98 |
| chr1_240161229_240161618 | 0.12 |
FMN2 |
formin 2 |
16225 |
0.27 |
| chr6_10418997_10419238 | 0.12 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
754 |
0.63 |
| chr15_25199832_25199983 | 0.12 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
167 |
0.55 |
| chr5_139050794_139051015 | 0.12 |
CXXC5 |
CXXC finger protein 5 |
4117 |
0.25 |
| chr16_1458660_1458855 | 0.12 |
LA16c-312E8.2 |
|
78 |
0.93 |
| chr11_111794191_111794413 | 0.12 |
CRYAB |
crystallin, alpha B |
144 |
0.91 |
| chr17_76879212_76879961 | 0.12 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9354 |
0.14 |
| chr9_139429643_139429794 | 0.12 |
RP11-413M3.4 |
|
7615 |
0.1 |
| chr19_10399295_10399446 | 0.12 |
CTD-2369P2.8 |
|
325 |
0.66 |
| chr1_1292304_1292633 | 0.12 |
MXRA8 |
matrix-remodelling associated 8 |
1447 |
0.18 |
| chr11_1329925_1330473 | 0.12 |
TOLLIP |
toll interacting protein |
650 |
0.47 |
| chr1_208137449_208137600 | 0.12 |
CD34 |
CD34 molecule |
52777 |
0.17 |
| chr19_1154555_1155129 | 0.12 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr6_85472288_85472456 | 0.12 |
TBX18 |
T-box 18 |
701 |
0.8 |
| chr5_14204410_14204680 | 0.12 |
TRIO |
trio Rho guanine nucleotide exchange factor |
20638 |
0.29 |
| chr7_30028817_30029270 | 0.12 |
SCRN1 |
secernin 1 |
285 |
0.79 |
| chr11_75210907_75211158 | 0.11 |
RP11-939C17.4 |
|
8404 |
0.15 |
| chr9_72658914_72659269 | 0.11 |
MAMDC2 |
MAM domain containing 2 |
594 |
0.81 |
| chr10_29922441_29923089 | 0.11 |
SVIL |
supervillin |
1136 |
0.54 |
| chr11_503866_504897 | 0.11 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
171 |
0.89 |
| chrX_51239510_51239840 | 0.11 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
227 |
0.96 |
| chr7_64700247_64700513 | 0.11 |
ENSG00000265175 |
. |
27929 |
0.19 |
| chr12_112560647_112560960 | 0.11 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
2502 |
0.22 |
| chr10_104359700_104359851 | 0.11 |
ENSG00000207029 |
. |
8494 |
0.17 |
| chr1_227506323_227506932 | 0.11 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
452 |
0.9 |
| chr17_77756152_77756303 | 0.11 |
CBX2 |
chromobox homolog 2 |
4234 |
0.16 |
| chr12_105477204_105477480 | 0.11 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
999 |
0.49 |
| chr4_177712658_177713139 | 0.11 |
VEGFC |
vascular endothelial growth factor C |
983 |
0.71 |
| chr21_45553858_45554178 | 0.11 |
C21orf33 |
chromosome 21 open reading frame 33 |
322 |
0.87 |
| chr17_39970124_39970275 | 0.11 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
1016 |
0.29 |
| chr7_5467738_5468320 | 0.11 |
TNRC18 |
trinucleotide repeat containing 18 |
2984 |
0.19 |
| chr4_8272289_8272466 | 0.11 |
HTRA3 |
HtrA serine peptidase 3 |
873 |
0.65 |
| chr11_14403032_14403213 | 0.11 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
17070 |
0.25 |
| chr6_21665058_21665374 | 0.11 |
SOX4 |
SRY (sex determining region Y)-box 4 |
71145 |
0.14 |
| chr14_103058041_103058305 | 0.11 |
RCOR1 |
REST corepressor 1 |
825 |
0.6 |
| chr3_46972745_46972896 | 0.11 |
PTH1R |
parathyroid hormone 1 receptor |
28731 |
0.12 |
| chr7_27191813_27192140 | 0.11 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
2305 |
0.1 |
| chr20_4878730_4878881 | 0.11 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
1488 |
0.39 |
| chr17_15850937_15851088 | 0.11 |
ADORA2B |
adenosine A2b receptor |
2781 |
0.26 |
| chr3_32001454_32001950 | 0.11 |
OSBPL10 |
oxysterol binding protein-like 10 |
21090 |
0.19 |
| chr17_15870184_15870534 | 0.11 |
ADORA2B |
adenosine A2b receptor |
22128 |
0.14 |
| chr6_143014333_143014633 | 0.11 |
RP1-67K17.3 |
|
55097 |
0.17 |
| chr8_18942225_18942376 | 0.11 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
60 |
0.99 |
| chr7_28724839_28725039 | 0.11 |
CREB5 |
cAMP responsive element binding protein 5 |
659 |
0.83 |
| chr1_206255126_206255277 | 0.11 |
ENSG00000252692 |
. |
5892 |
0.18 |
| chr18_67069113_67069341 | 0.11 |
DOK6 |
docking protein 6 |
936 |
0.74 |
| chr9_38067740_38068285 | 0.11 |
SHB |
Src homology 2 domain containing adaptor protein B |
1196 |
0.58 |
| chr2_238813531_238813682 | 0.11 |
ENSG00000263723 |
. |
35059 |
0.15 |
| chr17_12877207_12877393 | 0.11 |
ENSG00000266152 |
. |
1158 |
0.45 |
| chrX_129657730_129658601 | 0.11 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
122186 |
0.05 |
| chr2_120189452_120190154 | 0.11 |
TMEM37 |
transmembrane protein 37 |
358 |
0.88 |
| chr10_49678072_49678223 | 0.11 |
ARHGAP22 |
Rho GTPase activating protein 22 |
18477 |
0.21 |
| chr5_129240122_129240866 | 0.10 |
CHSY3 |
chondroitin sulfate synthase 3 |
329 |
0.79 |
| chr9_130659943_130660240 | 0.10 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
200 |
0.87 |
| chr19_13392901_13393052 | 0.10 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
6266 |
0.22 |
| chr5_150863261_150863412 | 0.10 |
ENSG00000200227 |
. |
6198 |
0.17 |
| chr11_17096518_17096669 | 0.10 |
ENSG00000272034 |
. |
302 |
0.67 |
| chr1_208133085_208133236 | 0.10 |
CD34 |
CD34 molecule |
48413 |
0.18 |
| chr17_45277656_45277848 | 0.10 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
8649 |
0.12 |
| chr8_104152453_104152604 | 0.10 |
BAALC |
brain and acute leukemia, cytoplasmic |
410 |
0.69 |
| chr17_79859390_79859541 | 0.10 |
NPB |
neuropeptide B |
520 |
0.48 |
| chr4_75480156_75480665 | 0.10 |
AREGB |
amphiregulin B |
219 |
0.94 |
| chr6_166074004_166074283 | 0.10 |
PDE10A |
phosphodiesterase 10A |
1414 |
0.55 |
| chr15_81072509_81072719 | 0.10 |
KIAA1199 |
KIAA1199 |
902 |
0.65 |
| chr11_61595808_61596402 | 0.10 |
FADS2 |
fatty acid desaturase 2 |
353 |
0.64 |
| chr1_119521934_119522435 | 0.10 |
TBX15 |
T-box 15 |
8244 |
0.28 |
| chr17_2613659_2614081 | 0.10 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
622 |
0.56 |
| chr9_90131995_90132200 | 0.10 |
DAPK1 |
death-associated protein kinase 1 |
18177 |
0.23 |
| chr9_130308551_130308785 | 0.10 |
FAM129B |
family with sequence similarity 129, member B |
22699 |
0.15 |
| chr10_95225832_95226004 | 0.10 |
MYOF |
myoferlin |
16033 |
0.19 |
| chr1_16087111_16087637 | 0.10 |
FBLIM1 |
filamin binding LIM protein 1 |
2082 |
0.2 |
| chr4_75310406_75310883 | 0.10 |
AREG |
amphiregulin |
207 |
0.94 |
| chr4_90757689_90758348 | 0.10 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
109 |
0.84 |
| chr10_99344095_99344388 | 0.10 |
PI4K2A |
Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
110 |
0.53 |
| chr19_47991140_47991431 | 0.10 |
NAPA-AS1 |
NAPA antisense RNA 1 |
3720 |
0.15 |
| chr3_38325709_38325860 | 0.10 |
SLC22A14 |
solute carrier family 22, member 14 |
1995 |
0.29 |
| chr3_184286341_184286629 | 0.10 |
EPHB3 |
EPH receptor B3 |
6913 |
0.2 |
| chr16_2204070_2204407 | 0.10 |
ENSG00000206630 |
. |
868 |
0.23 |
| chr4_7907648_7907905 | 0.10 |
AC097381.1 |
|
32952 |
0.16 |
| chr22_20790240_20790391 | 0.10 |
SCARF2 |
scavenger receptor class F, member 2 |
1797 |
0.22 |
| chr2_132430511_132430704 | 0.10 |
C2orf27A |
chromosome 2 open reading frame 27A |
49341 |
0.15 |
| chr9_130640034_130640202 | 0.10 |
AK1 |
adenylate kinase 1 |
96 |
0.92 |
| chr19_13734648_13734872 | 0.10 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
44 |
0.98 |
| chr5_52105955_52106201 | 0.10 |
CTD-2288O8.1 |
|
22218 |
0.18 |
| chr1_84158363_84158987 | 0.10 |
ENSG00000223231 |
. |
100885 |
0.09 |
| chr1_157963513_157964097 | 0.09 |
KIRREL |
kin of IRRE like (Drosophila) |
370 |
0.89 |
| chr6_158478262_158478415 | 0.09 |
SYNJ2 |
synaptojanin 2 |
20374 |
0.19 |
| chr12_49183193_49183344 | 0.09 |
ADCY6 |
adenylate cyclase 6 |
448 |
0.69 |
| chr4_170192362_170192994 | 0.09 |
SH3RF1 |
SH3 domain containing ring finger 1 |
422 |
0.9 |
| chr7_11579827_11580194 | 0.09 |
AC004160.4 |
|
133428 |
0.05 |
| chr10_79470838_79471281 | 0.09 |
ENSG00000199664 |
. |
65754 |
0.1 |
| chr5_79866210_79866442 | 0.09 |
ANKRD34B |
ankyrin repeat domain 34B |
19 |
0.98 |
| chr9_137236302_137236526 | 0.09 |
RXRA |
retinoid X receptor, alpha |
17988 |
0.22 |
| chr10_43573048_43573466 | 0.09 |
RET |
ret proto-oncogene |
730 |
0.74 |
| chr20_50417404_50418104 | 0.09 |
SALL4 |
spalt-like transcription factor 4 |
1193 |
0.55 |
| chr19_47522872_47523023 | 0.09 |
NPAS1 |
neuronal PAS domain protein 1 |
130 |
0.96 |
| chr19_18521360_18521522 | 0.09 |
ENSG00000221167 |
. |
849 |
0.39 |
| chr13_110959773_110960124 | 0.09 |
COL4A2 |
collagen, type IV, alpha 2 |
334 |
0.6 |
| chr13_44730017_44730207 | 0.09 |
SMIM2-IT1 |
SMIM2 intronic transcript 1 (non-protein coding) |
2246 |
0.3 |
| chr1_1291746_1291897 | 0.09 |
MXRA8 |
matrix-remodelling associated 8 |
2094 |
0.12 |
| chr2_10540557_10540762 | 0.09 |
HPCAL1 |
hippocalcin-like 1 |
19488 |
0.18 |
| chr8_687260_687856 | 0.09 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
93 |
0.97 |
| chr19_45146992_45147170 | 0.09 |
PVR |
poliovirus receptor |
17 |
0.97 |
| chr17_76921771_76922019 | 0.09 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
426 |
0.79 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0071605 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.3 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.2 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |