Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
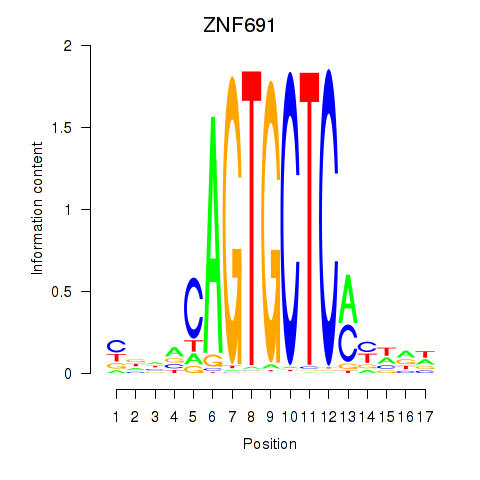
Results for ZNF691
Z-value: 1.14
Transcription factors associated with ZNF691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF691
|
ENSG00000164011.13 | zinc finger protein 691 |
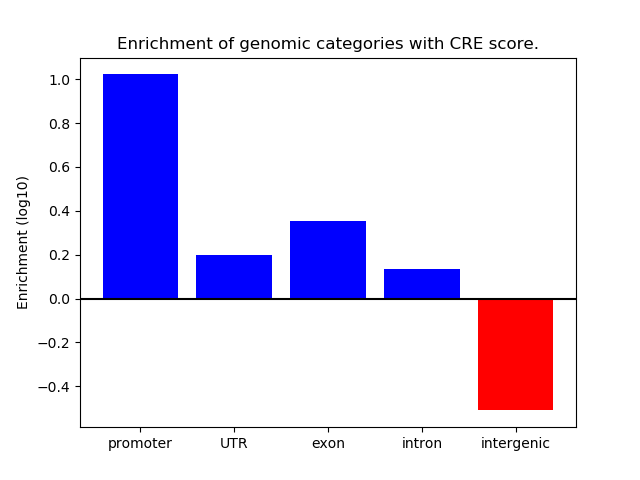
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_43316561_43316712 | ZNF691 | 278 | 0.868747 | -0.76 | 1.7e-02 | Click! |
| chr1_43314081_43314232 | ZNF691 | 806 | 0.493229 | 0.54 | 1.4e-01 | Click! |
| chr1_43312221_43312990 | ZNF691 | 297 | 0.646867 | 0.03 | 9.3e-01 | Click! |
Activity of the ZNF691 motif across conditions
Conditions sorted by the z-value of the ZNF691 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
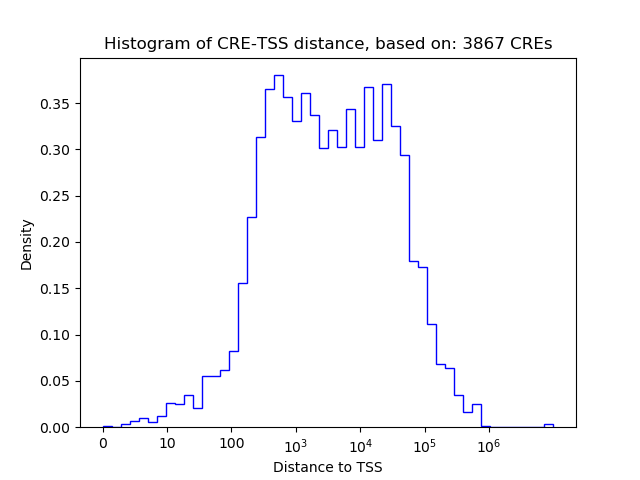
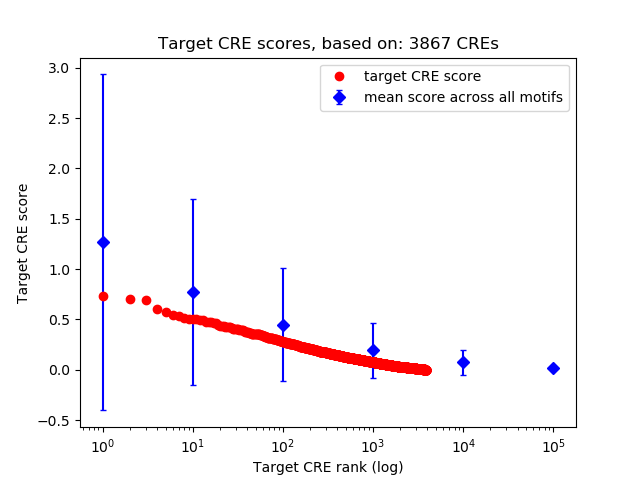
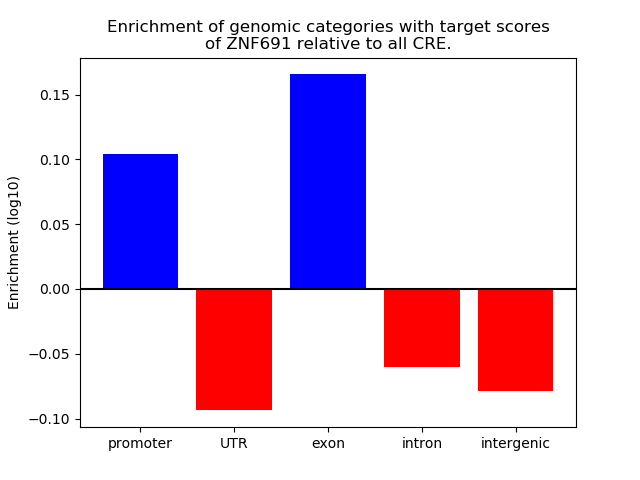
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_50523508_50523892 | 0.74 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
12 |
0.97 |
| chr3_56950582_56950733 | 0.71 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
158 |
0.97 |
| chr10_9058777_9059039 | 0.70 |
ENSG00000212505 |
. |
360114 |
0.01 |
| chr14_22971510_22971661 | 0.60 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
15414 |
0.1 |
| chr14_23000842_23001170 | 0.58 |
TRAJ15 |
T cell receptor alpha joining 15 |
2426 |
0.17 |
| chr12_5996777_5996928 | 0.55 |
ANO2 |
anoctamin 2 |
58546 |
0.15 |
| chr4_108971019_108971228 | 0.53 |
HADH |
hydroxyacyl-CoA dehydrogenase |
45332 |
0.15 |
| chr6_33044279_33044526 | 0.52 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
659 |
0.57 |
| chr3_196367146_196367308 | 0.51 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
581 |
0.45 |
| chr1_111739082_111739233 | 0.50 |
DENND2D |
DENN/MADD domain containing 2D |
4154 |
0.14 |
| chr15_81592474_81592729 | 0.50 |
IL16 |
interleukin 16 |
844 |
0.63 |
| chr3_112216938_112217091 | 0.50 |
BTLA |
B and T lymphocyte associated |
1191 |
0.55 |
| chr13_24826937_24827088 | 0.49 |
SPATA13 |
spermatogenesis associated 13 |
1156 |
0.4 |
| chr1_198628489_198628711 | 0.48 |
RP11-553K8.5 |
|
7590 |
0.25 |
| chr1_111213979_111214130 | 0.47 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
3601 |
0.23 |
| chr7_142494445_142494858 | 0.47 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
13520 |
0.18 |
| chr14_22539203_22539554 | 0.47 |
ENSG00000238634 |
. |
71509 |
0.12 |
| chr3_156192769_156192920 | 0.46 |
KCNAB1-AS1 |
KCNAB1 antisense RNA 1 |
28150 |
0.2 |
| chr13_30948639_30948822 | 0.44 |
KATNAL1 |
katanin p60 subunit A-like 1 |
67109 |
0.12 |
| chr14_102293036_102293187 | 0.44 |
CTD-2017C7.1 |
|
12757 |
0.15 |
| chr19_18230369_18230520 | 0.44 |
IL12RB1 |
interleukin 12 receptor, beta 1 |
20690 |
0.1 |
| chr3_186743992_186744192 | 0.43 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
821 |
0.7 |
| chr2_74518166_74518317 | 0.43 |
SLC4A5 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
409 |
0.81 |
| chr1_226067206_226067421 | 0.43 |
TMEM63A |
transmembrane protein 63A |
1934 |
0.23 |
| chr5_133455627_133455909 | 0.43 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3541 |
0.27 |
| chr19_51873675_51874043 | 0.43 |
CLDND2 |
claudin domain containing 2 |
1602 |
0.15 |
| chr14_22970808_22971009 | 0.42 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
14737 |
0.1 |
| chr6_112078365_112078719 | 0.41 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1775 |
0.47 |
| chr10_72192454_72192856 | 0.41 |
AC022532.1 |
Uncharacterized protein |
1930 |
0.29 |
| chr11_65321714_65321865 | 0.40 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
591 |
0.53 |
| chr2_175456506_175456946 | 0.40 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
5767 |
0.19 |
| chr6_170405095_170405246 | 0.40 |
RP11-302L19.1 |
|
72571 |
0.11 |
| chr13_34184837_34185306 | 0.40 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
207115 |
0.02 |
| chr8_134069456_134069607 | 0.39 |
SLA |
Src-like-adaptor |
3072 |
0.32 |
| chr22_23799075_23799226 | 0.39 |
ZDHHC8P1 |
zinc finger, DHHC-type containing 8 pseudogene 1 |
54806 |
0.11 |
| chr13_41553204_41553401 | 0.39 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
3116 |
0.27 |
| chr6_35016533_35016804 | 0.38 |
TCP11 |
t-complex 11, testis-specific |
72154 |
0.1 |
| chr6_41048535_41048765 | 0.38 |
NFYA |
nuclear transcription factor Y, alpha |
7928 |
0.13 |
| chr3_56950038_56950370 | 0.38 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
295 |
0.93 |
| chr6_108144289_108144729 | 0.37 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
1007 |
0.66 |
| chr14_22947996_22948147 | 0.37 |
ENSG00000251002 |
. |
2207 |
0.16 |
| chr2_223290336_223290619 | 0.37 |
SGPP2 |
sphingosine-1-phosphate phosphatase 2 |
1241 |
0.5 |
| chr12_9911079_9911493 | 0.36 |
CD69 |
CD69 molecule |
2211 |
0.26 |
| chr18_13216452_13216669 | 0.36 |
RP11-794M8.1 |
|
194 |
0.92 |
| chr6_139467434_139467700 | 0.36 |
HECA |
headcase homolog (Drosophila) |
11318 |
0.25 |
| chr11_65883776_65883927 | 0.36 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
15617 |
0.11 |
| chr14_23007518_23007669 | 0.36 |
TRAJ15 |
T cell receptor alpha joining 15 |
9013 |
0.11 |
| chr2_204733780_204733985 | 0.36 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
1073 |
0.62 |
| chr13_74706686_74707383 | 0.36 |
KLF12 |
Kruppel-like factor 12 |
1360 |
0.61 |
| chr22_42177525_42177912 | 0.36 |
MEI1 |
meiosis inhibitor 1 |
21 |
0.96 |
| chr14_22772288_22772466 | 0.36 |
ENSG00000251002 |
. |
129342 |
0.04 |
| chr13_100309364_100309515 | 0.36 |
ENSG00000263615 |
. |
14126 |
0.18 |
| chr17_29135904_29136074 | 0.35 |
CRLF3 |
cytokine receptor-like factor 3 |
15700 |
0.12 |
| chr20_39769151_39769427 | 0.35 |
RP1-1J6.2 |
|
2646 |
0.27 |
| chr16_27241764_27241915 | 0.35 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
2560 |
0.25 |
| chr2_46578889_46579334 | 0.35 |
EPAS1 |
endothelial PAS domain protein 1 |
54570 |
0.13 |
| chr12_51716594_51716774 | 0.35 |
BIN2 |
bridging integrator 2 |
1215 |
0.42 |
| chr18_60823743_60823894 | 0.35 |
RP11-299P2.1 |
|
5265 |
0.26 |
| chr10_98134778_98135198 | 0.34 |
OPALIN |
oligodendrocytic myelin paranodal and inner loop protein |
15896 |
0.21 |
| chr2_10522774_10523051 | 0.34 |
HPCAL1 |
hippocalcin-like 1 |
37235 |
0.15 |
| chr17_41738444_41738624 | 0.33 |
MEOX1 |
mesenchyme homeobox 1 |
397 |
0.85 |
| chr7_7960072_7960471 | 0.33 |
ENSG00000201747 |
. |
14121 |
0.18 |
| chr9_116911638_116911789 | 0.33 |
COL27A1 |
collagen, type XXVII, alpha 1 |
6127 |
0.23 |
| chr2_68961577_68961872 | 0.33 |
ARHGAP25 |
Rho GTPase activating protein 25 |
189 |
0.96 |
| chr7_21395262_21395449 | 0.33 |
ENSG00000195024 |
. |
25626 |
0.21 |
| chr12_55378635_55378823 | 0.32 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
199 |
0.95 |
| chr12_92933741_92933892 | 0.32 |
ENSG00000238865 |
. |
4947 |
0.3 |
| chr16_88522781_88522932 | 0.32 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
3131 |
0.22 |
| chr15_26104694_26104860 | 0.32 |
ATP10A |
ATPase, class V, type 10A |
3578 |
0.24 |
| chr6_62995563_62995714 | 0.32 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
494 |
0.9 |
| chr11_65647291_65647461 | 0.31 |
CTSW |
cathepsin W |
47 |
0.93 |
| chr18_8441840_8441991 | 0.31 |
ENSG00000242985 |
. |
29914 |
0.14 |
| chr2_135464562_135464727 | 0.31 |
TMEM163 |
transmembrane protein 163 |
11926 |
0.28 |
| chr15_40600356_40600529 | 0.31 |
PLCB2 |
phospholipase C, beta 2 |
319 |
0.77 |
| chr9_130740055_130740787 | 0.31 |
FAM102A |
family with sequence similarity 102, member A |
2371 |
0.18 |
| chr17_48047829_48047980 | 0.31 |
DLX4 |
distal-less homeobox 4 |
1210 |
0.4 |
| chr13_40397646_40397797 | 0.31 |
ENSG00000212553 |
. |
33643 |
0.19 |
| chr8_111604369_111604520 | 0.31 |
KCNV1 |
potassium channel, subfamily V, member 1 |
616368 |
0.0 |
| chr22_36425686_36425854 | 0.31 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
1297 |
0.58 |
| chr5_179235168_179235459 | 0.31 |
MGAT4B |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
1379 |
0.24 |
| chr22_27843716_27843867 | 0.31 |
RP11-375H17.1 |
|
268677 |
0.02 |
| chr2_33432670_33432821 | 0.31 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
35984 |
0.2 |
| chr20_44834342_44834493 | 0.31 |
CDH22 |
cadherin 22, type 2 |
45917 |
0.14 |
| chr10_9271793_9271944 | 0.30 |
ENSG00000212505 |
. |
573074 |
0.0 |
| chr8_82044855_82045010 | 0.30 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
20629 |
0.26 |
| chr13_100072782_100072933 | 0.30 |
ENSG00000266207 |
. |
32866 |
0.16 |
| chr12_739807_740337 | 0.30 |
RP11-218M22.1 |
|
15 |
0.97 |
| chr3_50487862_50488065 | 0.30 |
ENSG00000202322 |
. |
30751 |
0.11 |
| chr5_133772510_133772733 | 0.30 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
25032 |
0.14 |
| chr3_60061066_60061217 | 0.29 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
103558 |
0.09 |
| chrX_12992472_12992678 | 0.29 |
TMSB4X |
thymosin beta 4, X-linked |
652 |
0.76 |
| chr2_231523885_231524103 | 0.29 |
CAB39 |
calcium binding protein 39 |
53566 |
0.12 |
| chr2_43229390_43229541 | 0.29 |
ENSG00000207087 |
. |
89167 |
0.09 |
| chr1_167598231_167598382 | 0.29 |
RCSD1 |
RCSD domain containing 1 |
1024 |
0.47 |
| chr5_169729941_169730225 | 0.29 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4852 |
0.25 |
| chr1_40500456_40500611 | 0.29 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
5372 |
0.21 |
| chr9_100667248_100667478 | 0.28 |
C9orf156 |
chromosome 9 open reading frame 156 |
7390 |
0.17 |
| chr9_136003459_136003610 | 0.28 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
1254 |
0.38 |
| chr20_4794500_4794911 | 0.28 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
1064 |
0.55 |
| chr20_57721382_57721625 | 0.28 |
ZNF831 |
zinc finger protein 831 |
44572 |
0.15 |
| chr21_32534825_32534976 | 0.28 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
32361 |
0.22 |
| chr2_10472002_10472153 | 0.28 |
HPCAL1 |
hippocalcin-like 1 |
28251 |
0.15 |
| chr12_42863299_42863450 | 0.28 |
RP11-328C8.4 |
|
10206 |
0.15 |
| chr10_90911880_90912031 | 0.27 |
CH25H |
cholesterol 25-hydroxylase |
55116 |
0.1 |
| chr17_4618141_4618477 | 0.27 |
ARRB2 |
arrestin, beta 2 |
575 |
0.53 |
| chrX_18359237_18359476 | 0.27 |
SCML2 |
sex comb on midleg-like 2 (Drosophila) |
13491 |
0.24 |
| chr11_128344340_128344491 | 0.27 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
30874 |
0.21 |
| chr12_56881156_56881307 | 0.27 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
671 |
0.58 |
| chr15_85711869_85712366 | 0.27 |
CSPG4P12 |
chondroitin sulfate proteoglycan 4 pseudogene 12 |
22552 |
0.15 |
| chr16_68114330_68114481 | 0.27 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
4842 |
0.12 |
| chrX_107068956_107069440 | 0.27 |
MID2 |
midline 2 |
89 |
0.97 |
| chr1_150536239_150536708 | 0.27 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
2504 |
0.12 |
| chr11_104914233_104914622 | 0.27 |
CARD16 |
caspase recruitment domain family, member 16 |
1607 |
0.35 |
| chr18_75362846_75362997 | 0.27 |
ENSG00000252260 |
. |
212602 |
0.02 |
| chr3_113954111_113954262 | 0.27 |
ZNF80 |
zinc finger protein 80 |
2239 |
0.27 |
| chr8_134511475_134511742 | 0.26 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
4 |
0.99 |
| chr18_74766190_74766781 | 0.26 |
MBP |
myelin basic protein |
36751 |
0.2 |
| chr6_166747013_166747164 | 0.26 |
SFT2D1 |
SFT2 domain containing 1 |
8991 |
0.19 |
| chr18_2636542_2636784 | 0.26 |
ENSG00000200875 |
. |
13203 |
0.14 |
| chr3_45732221_45732372 | 0.26 |
SACM1L |
SAC1 suppressor of actin mutations 1-like (yeast) |
1345 |
0.36 |
| chrX_153166301_153166452 | 0.26 |
AVPR2 |
arginine vasopressin receptor 2 |
3813 |
0.11 |
| chrX_74742629_74743232 | 0.26 |
ZDHHC15 |
zinc finger, DHHC-type containing 15 |
57 |
0.99 |
| chr22_50318944_50319095 | 0.26 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
6638 |
0.17 |
| chr12_47605032_47605192 | 0.26 |
PCED1B |
PC-esterase domain containing 1B |
4940 |
0.24 |
| chr4_187407754_187407905 | 0.26 |
F11-AS1 |
F11 antisense RNA 1 |
14322 |
0.19 |
| chr19_10304500_10305008 | 0.26 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
548 |
0.62 |
| chr6_119027020_119027704 | 0.26 |
CEP85L |
centrosomal protein 85kDa-like |
3869 |
0.35 |
| chr3_9993293_9993483 | 0.26 |
PRRT3 |
proline-rich transmembrane protein 3 |
686 |
0.44 |
| chrX_68310760_68310911 | 0.25 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
71702 |
0.13 |
| chr16_29626573_29626859 | 0.25 |
ENSG00000266758 |
. |
16130 |
0.13 |
| chr22_39854364_39854515 | 0.25 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
1090 |
0.44 |
| chr15_60771714_60772006 | 0.25 |
RP11-219B17.1 |
|
441 |
0.52 |
| chr10_88299439_88299590 | 0.25 |
RP11-77P6.2 |
|
17812 |
0.18 |
| chr17_56412846_56413076 | 0.25 |
ENSG00000264399 |
. |
422 |
0.69 |
| chr6_32912081_32912232 | 0.25 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
3309 |
0.12 |
| chr3_186237256_186237407 | 0.25 |
CRYGS |
crystallin, gamma S |
24909 |
0.16 |
| chr5_79547739_79547890 | 0.25 |
SERINC5 |
serine incorporator 5 |
4024 |
0.22 |
| chr20_35258352_35258894 | 0.25 |
SLA2 |
Src-like-adaptor 2 |
15661 |
0.12 |
| chr15_63894184_63894335 | 0.25 |
USP3-AS1 |
USP3 antisense RNA 1 |
471 |
0.81 |
| chr4_38511545_38511706 | 0.25 |
RP11-617D20.1 |
|
114571 |
0.06 |
| chr9_20398478_20398629 | 0.24 |
ENSG00000263790 |
. |
12683 |
0.18 |
| chr14_99723060_99723211 | 0.24 |
AL109767.1 |
|
6150 |
0.23 |
| chr13_25172075_25172226 | 0.24 |
ENSG00000211508 |
. |
11067 |
0.18 |
| chr17_32576011_32576162 | 0.24 |
CCL2 |
chemokine (C-C motif) ligand 2 |
6218 |
0.14 |
| chr1_53103098_53103249 | 0.24 |
FAM159A |
family with sequence similarity 159, member A |
4157 |
0.21 |
| chr13_31481253_31481536 | 0.24 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
551 |
0.78 |
| chr4_150608043_150608194 | 0.24 |
ENSG00000202331 |
. |
220594 |
0.02 |
| chr13_49875518_49875737 | 0.24 |
CAB39L |
calcium binding protein 39-like |
49636 |
0.13 |
| chr11_44748940_44749566 | 0.24 |
TSPAN18 |
tetraspanin 18 |
529 |
0.86 |
| chr4_143328016_143328342 | 0.23 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
24233 |
0.29 |
| chr9_130212354_130212505 | 0.23 |
RPL12 |
ribosomal protein L12 |
1223 |
0.25 |
| chr7_115116722_115116873 | 0.23 |
ENSG00000202377 |
. |
104699 |
0.08 |
| chr12_120689652_120689816 | 0.23 |
PXN |
paxillin |
1770 |
0.24 |
| chr3_14467300_14467558 | 0.23 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
6749 |
0.24 |
| chr14_103541034_103541185 | 0.23 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
17310 |
0.15 |
| chr9_130539046_130539317 | 0.23 |
SH2D3C |
SH2 domain containing 3C |
1839 |
0.17 |
| chr1_116694143_116694294 | 0.23 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
39842 |
0.18 |
| chr14_57283183_57283334 | 0.23 |
OTX2-AS1 |
OTX2 antisense RNA 1 (head to head) |
3249 |
0.2 |
| chr7_100224057_100224300 | 0.23 |
TFR2 |
transferrin receptor 2 |
6549 |
0.09 |
| chr10_6088158_6088309 | 0.23 |
IL2RA |
interleukin 2 receptor, alpha |
16020 |
0.13 |
| chr3_106580754_106580905 | 0.23 |
ENSG00000252626 |
. |
145586 |
0.05 |
| chr2_196977844_196978082 | 0.23 |
RP11-347P5.1 |
|
38024 |
0.15 |
| chr12_113416849_113417235 | 0.23 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
697 |
0.66 |
| chr15_78339087_78339263 | 0.23 |
ENSG00000221476 |
. |
8302 |
0.14 |
| chrX_55545684_55545835 | 0.22 |
USP51 |
ubiquitin specific peptidase 51 |
30124 |
0.17 |
| chr4_2486639_2486842 | 0.22 |
RNF4 |
ring finger protein 4 |
4798 |
0.2 |
| chr12_55385441_55385592 | 0.22 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
6986 |
0.23 |
| chr2_68962051_68962936 | 0.22 |
ARHGAP25 |
Rho GTPase activating protein 25 |
479 |
0.86 |
| chrX_47508576_47509160 | 0.22 |
ELK1 |
ELK1, member of ETS oncogene family |
1019 |
0.41 |
| chr1_25800454_25800605 | 0.22 |
TMEM57 |
transmembrane protein 57 |
43118 |
0.11 |
| chr4_29508375_29508526 | 0.22 |
ENSG00000252303 |
. |
3807 |
0.39 |
| chr19_17516946_17517123 | 0.22 |
MVB12A |
multivesicular body subunit 12A |
80 |
0.86 |
| chr16_58534963_58535658 | 0.22 |
NDRG4 |
NDRG family member 4 |
62 |
0.96 |
| chrX_135056749_135057023 | 0.22 |
MMGT1 |
membrane magnesium transporter 1 |
664 |
0.7 |
| chr1_235097634_235097790 | 0.22 |
ENSG00000239690 |
. |
57779 |
0.14 |
| chr5_1313580_1313801 | 0.22 |
ENSG00000263670 |
. |
4198 |
0.2 |
| chr10_49731081_49731274 | 0.22 |
ARHGAP22 |
Rho GTPase activating protein 22 |
1104 |
0.56 |
| chr7_36817405_36817556 | 0.22 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
53326 |
0.14 |
| chr3_186490709_186491008 | 0.22 |
ENSG00000207505 |
. |
1122 |
0.25 |
| chr2_30726606_30726757 | 0.22 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
56412 |
0.15 |
| chr11_58903611_58903772 | 0.22 |
FAM111A |
family with sequence similarity 111, member A |
6530 |
0.18 |
| chr19_36036123_36036368 | 0.22 |
AD000090.2 |
|
149 |
0.62 |
| chr7_151078177_151078328 | 0.22 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
12336 |
0.13 |
| chr1_44870677_44871051 | 0.22 |
RNF220 |
ring finger protein 220 |
2 |
0.98 |
| chr3_186748872_186749023 | 0.22 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
5676 |
0.26 |
| chr11_118311027_118311308 | 0.22 |
KMT2A |
lysine (K)-specific methyltransferase 2A |
3690 |
0.13 |
| chr18_77492609_77492760 | 0.22 |
RP11-567M16.5 |
|
29033 |
0.2 |
| chr5_10632325_10632658 | 0.22 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
4154 |
0.28 |
| chr8_69687686_69687837 | 0.21 |
ENSG00000239184 |
. |
79883 |
0.12 |
| chr1_206675169_206675320 | 0.21 |
RP11-534L20.5 |
|
2037 |
0.24 |
| chr7_84569129_84569450 | 0.21 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
181962 |
0.03 |
| chr20_30432784_30432935 | 0.21 |
FOXS1 |
forkhead box S1 |
561 |
0.67 |
| chr17_46060430_46060581 | 0.21 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
6417 |
0.1 |
| chr20_17945955_17946160 | 0.21 |
ENSG00000212232 |
. |
2468 |
0.21 |
| chr19_30158859_30159039 | 0.21 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
1168 |
0.55 |
| chr2_158289727_158289878 | 0.21 |
CYTIP |
cytohesin 1 interacting protein |
6124 |
0.23 |
| chr11_122597673_122598179 | 0.21 |
ENSG00000239079 |
. |
907 |
0.67 |
| chr22_48494042_48494263 | 0.21 |
ENSG00000266508 |
. |
176024 |
0.04 |
| chr6_130846468_130846619 | 0.21 |
ENSG00000202438 |
. |
48714 |
0.16 |
| chr6_166649375_166649526 | 0.21 |
ENSG00000201292 |
. |
26559 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.3 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 0.2 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.2 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.5 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 | 0.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.2 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0022616 | DNA strand elongation(GO:0022616) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.4 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.3 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |