Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
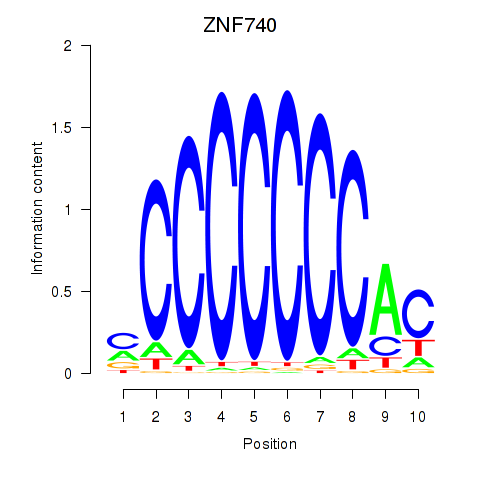
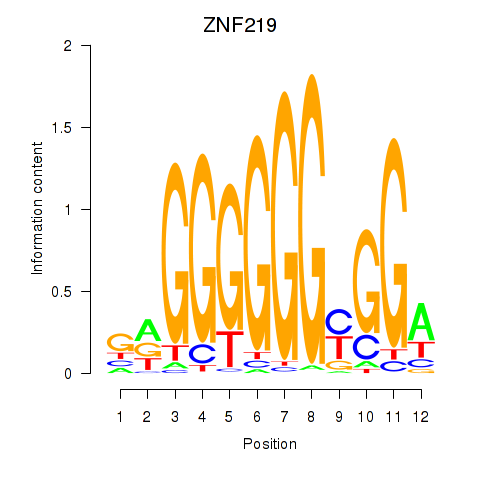
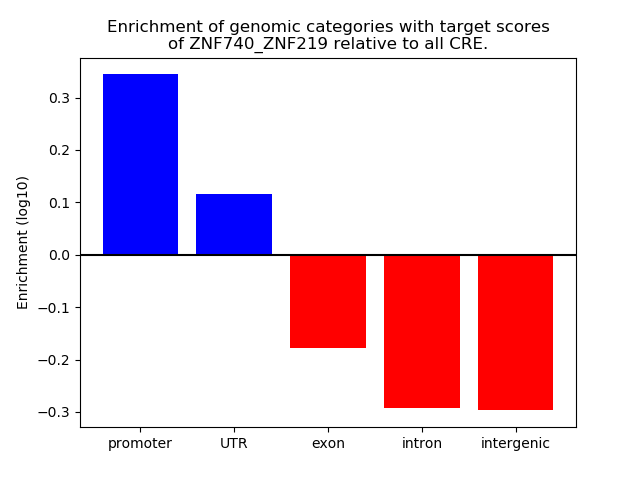
Results for ZNF740_ZNF219
Z-value: 1.09
Transcription factors associated with ZNF740_ZNF219
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF740
|
ENSG00000139651.9 | zinc finger protein 740 |
|
ZNF219
|
ENSG00000165804.11 | zinc finger protein 219 |
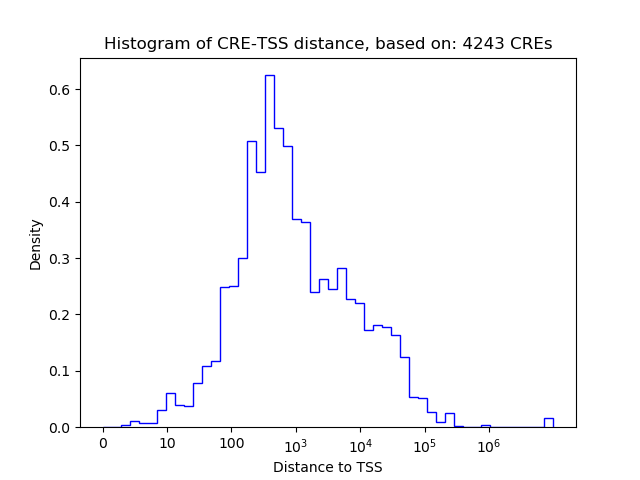
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_21554121_21554272 | ZNF219 | 7791 | 0.090077 | 0.72 | 3.0e-02 | Click! |
| chr14_21571987_21572281 | ZNF219 | 396 | 0.720913 | 0.66 | 5.5e-02 | Click! |
| chr14_21566135_21566286 | ZNF219 | 589 | 0.460361 | 0.62 | 7.4e-02 | Click! |
| chr14_21572301_21572831 | ZNF219 | 315 | 0.792366 | 0.62 | 7.5e-02 | Click! |
| chr14_21556081_21556232 | ZNF219 | 5831 | 0.096321 | 0.53 | 1.4e-01 | Click! |
| chr12_53574456_53574687 | ZNF740 | 87 | 0.539000 | 0.59 | 9.5e-02 | Click! |
Activity of the ZNF740_ZNF219 motif across conditions
Conditions sorted by the z-value of the ZNF740_ZNF219 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
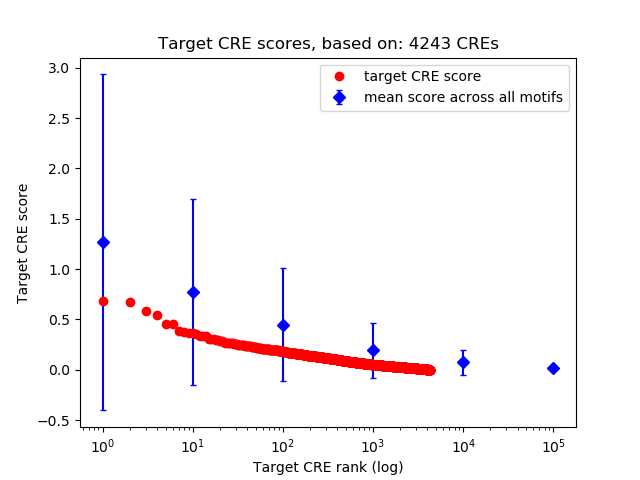
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_10341979_10342223 | 0.68 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
139 |
0.49 |
| chr11_47421960_47422248 | 0.67 |
ENSG00000264583 |
. |
417 |
0.68 |
| chr17_80176095_80176246 | 0.59 |
RP13-516M14.2 |
|
4067 |
0.12 |
| chr22_44577376_44577610 | 0.54 |
PARVG |
parvin, gamma |
215 |
0.96 |
| chr17_72743404_72743555 | 0.46 |
ENSG00000264624 |
. |
1273 |
0.21 |
| chr12_54694539_54694712 | 0.45 |
NFE2 |
nuclear factor, erythroid 2 |
174 |
0.67 |
| chr7_139761785_139761951 | 0.39 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
1653 |
0.43 |
| chr19_912362_912526 | 0.37 |
R3HDM4 |
R3H domain containing 4 |
787 |
0.39 |
| chr17_61773603_61773754 | 0.37 |
LIMD2 |
LIM domain containing 2 |
2844 |
0.21 |
| chr17_38478045_38478318 | 0.36 |
RARA |
retinoic acid receptor, alpha |
3644 |
0.15 |
| chr17_43300272_43300459 | 0.36 |
CTD-2020K17.1 |
|
776 |
0.38 |
| chr1_150540056_150540997 | 0.34 |
ENSG00000264508 |
. |
1190 |
0.22 |
| chr9_69785762_69786779 | 0.34 |
IGKV1OR-2 |
immunoglobulin kappa variable 1/OR-2 (pseudogene) |
8317 |
0.25 |
| chrX_40014744_40014994 | 0.33 |
BCOR |
BCL6 corepressor |
8987 |
0.31 |
| chr7_97910621_97910821 | 0.31 |
BRI3 |
brain protein I3 |
258 |
0.91 |
| chr1_40157482_40157633 | 0.30 |
HPCAL4 |
hippocalcin like 4 |
196 |
0.61 |
| chr3_107243151_107243484 | 0.30 |
BBX |
bobby sox homolog (Drosophila) |
113 |
0.98 |
| chr16_85647451_85647602 | 0.30 |
GSE1 |
Gse1 coiled-coil protein |
568 |
0.78 |
| chr1_32314203_32314496 | 0.29 |
SPOCD1 |
SPOC domain containing 1 |
32697 |
0.13 |
| chr19_14494334_14494521 | 0.28 |
CD97 |
CD97 molecule |
2171 |
0.24 |
| chr15_31656981_31657132 | 0.28 |
KLF13 |
Kruppel-like factor 13 |
1301 |
0.61 |
| chrX_48774424_48774575 | 0.27 |
PIM2 |
pim-2 oncogene |
1487 |
0.23 |
| chr14_52779713_52779864 | 0.27 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
1235 |
0.57 |
| chr11_129872420_129873004 | 0.27 |
PRDM10 |
PR domain containing 10 |
13 |
0.98 |
| chr19_52133397_52133630 | 0.27 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
75 |
0.95 |
| chr6_4777132_4777289 | 0.27 |
CDYL |
chromodomain protein, Y-like |
541 |
0.75 |
| chr19_4543528_4543679 | 0.26 |
ENSG00000263974 |
. |
1604 |
0.21 |
| chr2_157189811_157190046 | 0.26 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
696 |
0.78 |
| chr17_56405619_56405770 | 0.26 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
249 |
0.73 |
| chr7_21467616_21467934 | 0.26 |
SP4 |
Sp4 transcription factor |
114 |
0.97 |
| chr19_1068246_1068401 | 0.25 |
HMHA1 |
histocompatibility (minor) HA-1 |
826 |
0.4 |
| chr10_61659826_61659977 | 0.25 |
CCDC6 |
coiled-coil domain containing 6 |
6513 |
0.28 |
| chr18_77793817_77793982 | 0.25 |
TXNL4A |
thioredoxin-like 4A |
8 |
0.89 |
| chr22_20784096_20784540 | 0.25 |
SCARF2 |
scavenger receptor class F, member 2 |
7794 |
0.11 |
| chr2_54787203_54787366 | 0.25 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
1753 |
0.36 |
| chr21_44204010_44204161 | 0.25 |
AP001627.1 |
|
42217 |
0.15 |
| chrX_48542765_48543057 | 0.24 |
WAS |
Wiskott-Aldrich syndrome |
743 |
0.54 |
| chr17_76128459_76128696 | 0.24 |
TMC6 |
transmembrane channel-like 6 |
89 |
0.94 |
| chr12_4383631_4384229 | 0.24 |
CCND2-AS2 |
CCND2 antisense RNA 2 |
301 |
0.78 |
| chr11_1875039_1875307 | 0.24 |
LSP1 |
lymphocyte-specific protein 1 |
973 |
0.36 |
| chr6_137539516_137539806 | 0.24 |
IFNGR1 |
interferon gamma receptor 1 |
10 |
0.99 |
| chr9_136933745_136934121 | 0.23 |
BRD3 |
bromodomain containing 3 |
276 |
0.9 |
| chr17_40264582_40264792 | 0.23 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
45 |
0.95 |
| chr1_24969808_24970031 | 0.23 |
SRRM1 |
serine/arginine repetitive matrix 1 |
129 |
0.96 |
| chr12_122230893_122231238 | 0.23 |
RHOF |
ras homolog family member F (in filopodia) |
201 |
0.93 |
| chr2_233925739_233925978 | 0.23 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
669 |
0.7 |
| chr5_39201497_39202174 | 0.22 |
FYB |
FYN binding protein |
1294 |
0.59 |
| chr8_121823290_121823441 | 0.22 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
1018 |
0.42 |
| chr17_66192970_66193249 | 0.22 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
44500 |
0.11 |
| chr10_134368672_134368823 | 0.22 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
17104 |
0.21 |
| chr18_72922732_72923415 | 0.22 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
363 |
0.87 |
| chr12_31882551_31882882 | 0.22 |
AMN1 |
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
608 |
0.71 |
| chr5_131832293_131832540 | 0.22 |
IRF1 |
interferon regulatory factor 1 |
5926 |
0.15 |
| chr7_91569688_91570087 | 0.22 |
AKAP9 |
A kinase (PRKA) anchor protein 9 |
294 |
0.94 |
| chr19_4066716_4067152 | 0.22 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
9 |
0.96 |
| chrX_147691631_147691782 | 0.21 |
AFF2-IT1 |
AFF2 intronic transcript 1 (non-protein coding) |
63307 |
0.14 |
| chr2_43449640_43450246 | 0.21 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
3805 |
0.27 |
| chr1_236051645_236051799 | 0.21 |
LYST |
lysosomal trafficking regulator |
4850 |
0.19 |
| chr9_33817406_33817596 | 0.21 |
UBE2R2 |
ubiquitin-conjugating enzyme E2R 2 |
64 |
0.94 |
| chrX_2419173_2419324 | 0.21 |
DHRSX |
dehydrogenase/reductase (SDR family) X-linked |
229 |
0.51 |
| chr11_2326464_2326615 | 0.21 |
TSPAN32 |
tetraspanin 32 |
2441 |
0.18 |
| chr6_30798000_30798388 | 0.21 |
ENSG00000202241 |
. |
33833 |
0.07 |
| chrY_2369174_2369325 | 0.21 |
ENSG00000251841 |
. |
283541 |
0.01 |
| chr6_90793980_90794207 | 0.21 |
ENSG00000222078 |
. |
82868 |
0.09 |
| chr12_57870819_57871184 | 0.21 |
ARHGAP9 |
Rho GTPase activating protein 9 |
566 |
0.53 |
| chr10_82215935_82216086 | 0.20 |
TSPAN14 |
tetraspanin 14 |
1899 |
0.36 |
| chr19_12949779_12950060 | 0.20 |
MAST1 |
microtubule associated serine/threonine kinase 1 |
571 |
0.48 |
| chr12_94544055_94544362 | 0.20 |
PLXNC1 |
plexin C1 |
1709 |
0.42 |
| chr14_105527834_105528220 | 0.20 |
GPR132 |
G protein-coupled receptor 132 |
3740 |
0.23 |
| chrX_131157971_131158122 | 0.20 |
MST4 |
Serine/threonine-protein kinase MST4 |
410 |
0.87 |
| chr2_98547935_98548086 | 0.20 |
ENSG00000238719 |
. |
18430 |
0.21 |
| chr11_640222_640373 | 0.20 |
DRD4 |
dopamine receptor D4 |
3004 |
0.11 |
| chr14_99735608_99735969 | 0.20 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
1777 |
0.38 |
| chr1_27883616_27883836 | 0.20 |
RP1-159A19.4 |
|
31410 |
0.13 |
| chr1_156124352_156124692 | 0.20 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
360 |
0.78 |
| chr3_43329026_43329294 | 0.20 |
SNRK |
SNF related kinase |
1082 |
0.53 |
| chr6_1610062_1610262 | 0.20 |
FOXC1 |
forkhead box C1 |
519 |
0.87 |
| chr4_38665848_38666421 | 0.20 |
RP11-617D20.1 |
|
115 |
0.61 |
| chr7_949432_949583 | 0.20 |
ADAP1 |
ArfGAP with dual PH domains 1 |
4793 |
0.16 |
| chr12_51611544_51611860 | 0.20 |
POU6F1 |
POU class 6 homeobox 1 |
225 |
0.9 |
| chr18_60987754_60988433 | 0.20 |
BCL2 |
B-cell CLL/lymphoma 2 |
732 |
0.64 |
| chr17_29637369_29637894 | 0.20 |
EVI2B |
ecotropic viral integration site 2B |
3471 |
0.16 |
| chr19_3035801_3036232 | 0.19 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
6404 |
0.13 |
| chr15_69075318_69075469 | 0.19 |
ENSG00000265195 |
. |
18871 |
0.22 |
| chr17_21156722_21156881 | 0.19 |
C17orf103 |
chromosome 17 open reading frame 103 |
79 |
0.97 |
| chr19_16774664_16774815 | 0.19 |
TMEM38A |
transmembrane protein 38A |
2667 |
0.14 |
| chr17_57643262_57643540 | 0.19 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
417 |
0.85 |
| chr19_6481304_6482171 | 0.19 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr8_38759681_38760174 | 0.19 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
1065 |
0.52 |
| chr1_151431863_151432024 | 0.19 |
POGZ |
pogo transposable element with ZNF domain |
2 |
0.96 |
| chr11_441689_441900 | 0.19 |
ANO9 |
anoctamin 9 |
217 |
0.82 |
| chr18_60194123_60194717 | 0.19 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
3568 |
0.33 |
| chr12_124942372_124942523 | 0.19 |
NCOR2 |
nuclear receptor corepressor 2 |
25868 |
0.25 |
| chr17_55334331_55334657 | 0.19 |
MSI2 |
musashi RNA-binding protein 2 |
115 |
0.98 |
| chr20_48329140_48329431 | 0.19 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
1130 |
0.5 |
| chr1_26646074_26646278 | 0.19 |
UBXN11 |
UBX domain protein 11 |
1322 |
0.27 |
| chr7_45067326_45068016 | 0.18 |
CCM2 |
cerebral cavernous malformation 2 |
400 |
0.81 |
| chr17_4403063_4403461 | 0.18 |
AC118754.4 |
|
293 |
0.8 |
| chr19_42784997_42785148 | 0.18 |
CIC |
capicua transcriptional repressor |
3100 |
0.12 |
| chr19_47615950_47616307 | 0.18 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
403 |
0.69 |
| chr9_140348775_140349192 | 0.18 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
39 |
0.96 |
| chrX_12990058_12990209 | 0.18 |
TMSB4X |
thymosin beta 4, X-linked |
3094 |
0.3 |
| chr7_158656531_158656832 | 0.18 |
WDR60 |
WD repeat domain 60 |
5456 |
0.27 |
| chr13_74708290_74708441 | 0.18 |
KLF12 |
Kruppel-like factor 12 |
29 |
0.99 |
| chr15_78361308_78361764 | 0.18 |
TBC1D2B |
TBC1 domain family, member 2B |
3071 |
0.18 |
| chr15_41219932_41220083 | 0.18 |
DLL4 |
delta-like 4 (Drosophila) |
1584 |
0.27 |
| chr22_39323214_39323365 | 0.18 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
25467 |
0.12 |
| chr5_156696225_156696376 | 0.18 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
62 |
0.96 |
| chr15_100106167_100106396 | 0.18 |
MEF2A |
myocyte enhancer factor 2A |
11 |
0.98 |
| chr17_27139690_27139995 | 0.17 |
FAM222B |
family with sequence similarity 222, member B |
389 |
0.7 |
| chr1_21659698_21659849 | 0.17 |
ECE1 |
endothelin converting enzyme 1 |
12224 |
0.21 |
| chr15_65477311_65477678 | 0.17 |
CLPX |
caseinolytic mitochondrial matrix peptidase chaperone subunit |
186 |
0.93 |
| chr15_41953518_41954024 | 0.17 |
MGA |
MGA, MAX dimerization protein |
1161 |
0.47 |
| chr6_30649932_30650278 | 0.17 |
PPP1R18 |
protein phosphatase 1, regulatory subunit 18 |
4888 |
0.08 |
| chr14_74551356_74551507 | 0.17 |
LIN52 |
lin-52 homolog (C. elegans) |
68 |
0.72 |
| chr1_27816833_27817254 | 0.17 |
WASF2 |
WAS protein family, member 2 |
374 |
0.86 |
| chr9_20622216_20622410 | 0.17 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
201 |
0.95 |
| chr16_10710662_10710813 | 0.17 |
EMP2 |
epithelial membrane protein 2 |
36182 |
0.13 |
| chr11_2159500_2159786 | 0.17 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
561 |
0.59 |
| chr5_88589582_88589733 | 0.17 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
124967 |
0.06 |
| chr9_135114959_135115110 | 0.17 |
ENSG00000221105 |
. |
24772 |
0.21 |
| chr14_90084669_90085459 | 0.17 |
FOXN3 |
forkhead box N3 |
410 |
0.78 |
| chr11_108463883_108464034 | 0.16 |
EXPH5 |
exophilin 5 |
416 |
0.89 |
| chr17_2717574_2718059 | 0.16 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
18040 |
0.17 |
| chr2_43451286_43451513 | 0.16 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
2349 |
0.34 |
| chr19_15574852_15575181 | 0.16 |
RASAL3 |
RAS protein activator like 3 |
366 |
0.8 |
| chr19_17958305_17958511 | 0.16 |
JAK3 |
Janus kinase 3 |
418 |
0.74 |
| chr2_178129639_178129940 | 0.16 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
70 |
0.91 |
| chr7_129588935_129590397 | 0.16 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
1501 |
0.28 |
| chr8_145027093_145027557 | 0.16 |
PLEC |
plectin |
763 |
0.48 |
| chr10_3822916_3823697 | 0.16 |
KLF6 |
Kruppel-like factor 6 |
4079 |
0.25 |
| chr8_144108277_144108585 | 0.16 |
LY6E |
lymphocyte antigen 6 complex, locus E |
8457 |
0.17 |
| chr8_145639102_145639331 | 0.16 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
2680 |
0.09 |
| chr21_40759688_40760056 | 0.16 |
WRB |
tryptophan rich basic protein |
181 |
0.93 |
| chr2_237880489_237880640 | 0.16 |
ENSG00000202341 |
. |
43384 |
0.17 |
| chr14_24780359_24781199 | 0.16 |
LTB4R |
leukotriene B4 receptor |
74 |
0.58 |
| chr7_50345445_50345596 | 0.16 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1142 |
0.64 |
| chr2_85166549_85166831 | 0.16 |
KCMF1 |
potassium channel modulatory factor 1 |
31526 |
0.15 |
| chr7_1979405_1979706 | 0.16 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
630 |
0.8 |
| chr16_85558650_85558942 | 0.16 |
ENSG00000264203 |
. |
83698 |
0.08 |
| chr10_75633716_75633937 | 0.16 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
393 |
0.78 |
| chrX_39956550_39956704 | 0.16 |
BCOR |
BCL6 corepressor |
29 |
0.99 |
| chr19_912608_912979 | 0.16 |
R3HDM4 |
R3H domain containing 4 |
438 |
0.64 |
| chr11_66405551_66405805 | 0.16 |
RBM4 |
RNA binding motif protein 4 |
410 |
0.71 |
| chr22_32313508_32313842 | 0.16 |
C22orf24 |
chromosome 22 open reading frame 24 |
20728 |
0.14 |
| chr12_122243825_122244586 | 0.16 |
SETD1B |
SET domain containing 1B |
1567 |
0.32 |
| chr16_85646303_85646802 | 0.16 |
GSE1 |
Gse1 coiled-coil protein |
211 |
0.95 |
| chrY_1522891_1523042 | 0.16 |
NA |
NA |
> 106 |
NA |
| chr21_46348074_46348493 | 0.16 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
350 |
0.75 |
| chr3_194207533_194207696 | 0.16 |
ATP13A3 |
ATPase type 13A3 |
226 |
0.77 |
| chr9_130539046_130539317 | 0.16 |
SH2D3C |
SH2 domain containing 3C |
1839 |
0.17 |
| chr15_25684395_25684621 | 0.16 |
UBE3A |
ubiquitin protein ligase E3A |
380 |
0.82 |
| chr10_126849381_126849702 | 0.16 |
CTBP2 |
C-terminal binding protein 2 |
54 |
0.99 |
| chr16_3005959_3006166 | 0.15 |
LA16c-321D4.2 |
|
1785 |
0.16 |
| chr1_154976315_154976466 | 0.15 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
1095 |
0.25 |
| chr17_7787815_7787966 | 0.15 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
234 |
0.73 |
| chr17_32965108_32965375 | 0.15 |
TMEM132E |
transmembrane protein 132E |
57473 |
0.13 |
| chr11_71935004_71935259 | 0.15 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
143 |
0.92 |
| chr8_61591908_61592174 | 0.15 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
82 |
0.98 |
| chr19_14887538_14887792 | 0.15 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
11 |
0.97 |
| chr19_6728541_6728856 | 0.15 |
C3 |
complement component 3 |
1875 |
0.22 |
| chr4_185188898_185189263 | 0.15 |
ENSG00000221523 |
. |
496 |
0.82 |
| chr14_22958789_22958996 | 0.15 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
2721 |
0.15 |
| chr12_6716856_6717254 | 0.15 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
413 |
0.67 |
| chr16_85934507_85934793 | 0.15 |
IRF8 |
interferon regulatory factor 8 |
1645 |
0.44 |
| chr17_29820809_29821042 | 0.15 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
5799 |
0.15 |
| chr10_126840512_126840747 | 0.15 |
CTBP2 |
C-terminal binding protein 2 |
6656 |
0.3 |
| chr11_119598080_119598587 | 0.15 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
930 |
0.54 |
| chrX_70474174_70474507 | 0.15 |
ZMYM3 |
zinc finger, MYM-type 3 |
49 |
0.97 |
| chr7_73119920_73120071 | 0.15 |
ENSG00000265724 |
. |
5652 |
0.11 |
| chr20_4804308_4804651 | 0.15 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
188 |
0.95 |
| chr1_212781685_212782134 | 0.15 |
ATF3 |
activating transcription factor 3 |
103 |
0.96 |
| chr2_214006925_214007076 | 0.15 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
6353 |
0.32 |
| chr2_36683900_36684051 | 0.15 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
56858 |
0.13 |
| chr18_5295757_5296173 | 0.15 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
74 |
0.98 |
| chr19_10704173_10704324 | 0.15 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
6257 |
0.1 |
| chr10_3511482_3511923 | 0.15 |
RP11-184A2.3 |
|
281557 |
0.01 |
| chr4_78114724_78115069 | 0.15 |
CCNG2 |
cyclin G2 |
35317 |
0.18 |
| chr10_49893584_49893845 | 0.15 |
WDFY4 |
WDFY family member 4 |
793 |
0.69 |
| chr16_70473041_70473208 | 0.15 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
133 |
0.94 |
| chr1_156024285_156024442 | 0.15 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
180 |
0.82 |
| chr17_76124929_76125236 | 0.14 |
TMC6 |
transmembrane channel-like 6 |
219 |
0.87 |
| chr14_90863868_90864201 | 0.14 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
572 |
0.74 |
| chr20_30160458_30160983 | 0.14 |
HM13-AS1 |
HM13 antisense RNA 1 |
346 |
0.8 |
| chr12_27090458_27090857 | 0.14 |
ASUN |
asunder spermatogenesis regulator |
248 |
0.78 |
| chr19_45595083_45595471 | 0.14 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
941 |
0.32 |
| chr10_75632981_75633391 | 0.14 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
1033 |
0.4 |
| chr17_76127593_76127800 | 0.14 |
TMC6 |
transmembrane channel-like 6 |
792 |
0.35 |
| chr1_150551829_150551980 | 0.14 |
MCL1 |
myeloid cell leukemia sequence 1 (BCL2-related) |
102 |
0.92 |
| chr14_69259320_69259636 | 0.14 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
479 |
0.84 |
| chr1_2066282_2066433 | 0.14 |
PRKCZ |
protein kinase C, zeta |
72 |
0.96 |
| chr3_5231543_5231696 | 0.14 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
1585 |
0.26 |
| chr22_40390320_40390530 | 0.14 |
FAM83F |
family with sequence similarity 83, member F |
528 |
0.77 |
| chr9_139237156_139237308 | 0.14 |
GPSM1 |
G-protein signaling modulator 1 |
8327 |
0.12 |
| chr19_4610268_4610469 | 0.14 |
TNFAIP8L1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
29162 |
0.09 |
| chr19_53237403_53237997 | 0.14 |
ZNF611 |
zinc finger protein 611 |
597 |
0.71 |
| chr17_74268969_74269120 | 0.14 |
UBALD2 |
UBA-like domain containing 2 |
7260 |
0.13 |
| chr1_154975370_154975951 | 0.14 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
365 |
0.68 |
| chr3_136471001_136471256 | 0.14 |
STAG1 |
stromal antigen 1 |
80 |
0.81 |
| chrX_80064048_80064808 | 0.14 |
BRWD3 |
bromodomain and WD repeat domain containing 3 |
759 |
0.74 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.2 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.2 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0072193 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0090192 | regulation of glomerulus development(GO:0090192) |
| 0.0 | 0.1 | GO:0090504 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.3 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0043487 | regulation of RNA stability(GO:0043487) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0044766 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0003094 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.3 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 0.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.2 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.1 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0046033 | AMP metabolic process(GO:0046033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0035097 | methyltransferase complex(GO:0034708) histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |