Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
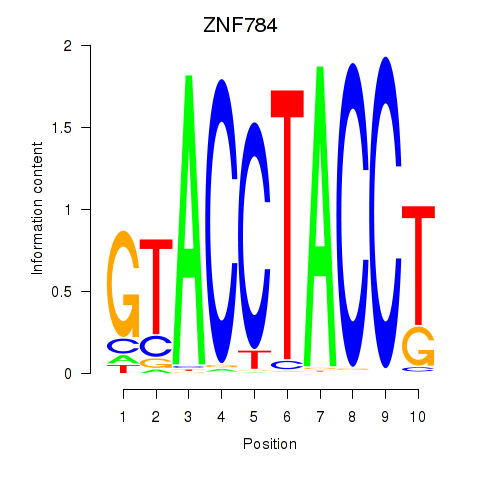
Results for ZNF784
Z-value: 1.72
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
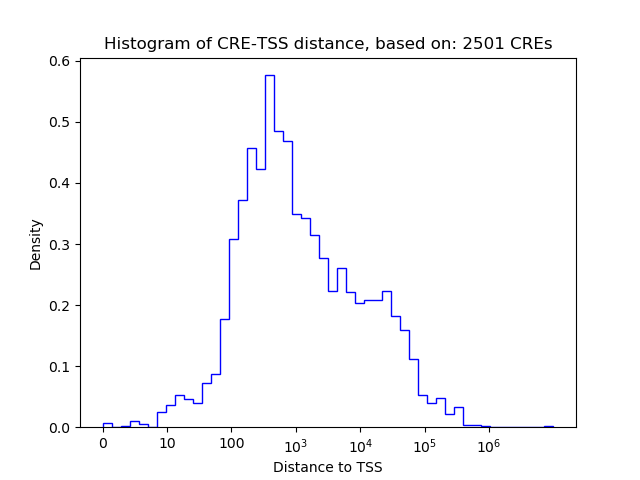
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_56136460_56136611 | ZNF784 | 568 | 0.492768 | 0.52 | 1.5e-01 | Click! |
| chr19_56132551_56132702 | ZNF784 | 3306 | 0.089935 | -0.44 | 2.3e-01 | Click! |
| chr19_56132152_56132303 | ZNF784 | 3705 | 0.085080 | -0.40 | 2.9e-01 | Click! |
| chr19_56135787_56135955 | ZNF784 | 61 | 0.922893 | 0.33 | 3.8e-01 | Click! |
| chr19_56135238_56135556 | ZNF784 | 535 | 0.519693 | -0.16 | 6.8e-01 | Click! |
Activity of the ZNF784 motif across conditions
Conditions sorted by the z-value of the ZNF784 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
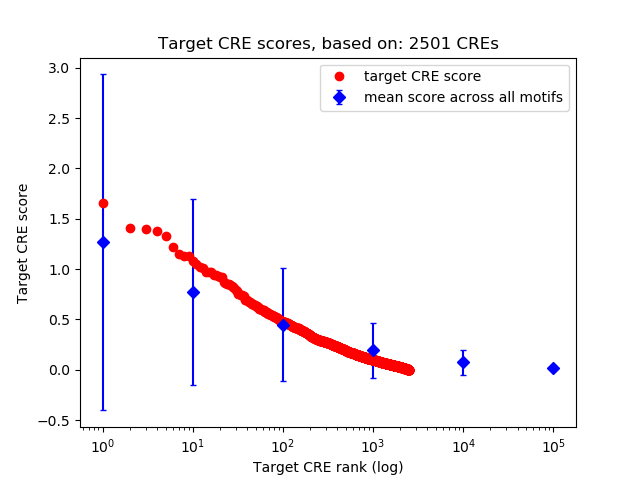
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_33864237_33864745 | 1.66 |
SLFN12L |
schlafen family member 12-like |
389 |
0.76 |
| chr3_121379556_121379755 | 1.40 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
90 |
0.96 |
| chr16_3116517_3116777 | 1.40 |
IL32 |
interleukin 32 |
834 |
0.3 |
| chr1_198608305_198608501 | 1.38 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
111 |
0.98 |
| chr10_5334923_5335219 | 1.33 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
4638 |
0.23 |
| chr1_207094900_207095227 | 1.22 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
149 |
0.94 |
| chr7_45067326_45068016 | 1.15 |
CCM2 |
cerebral cavernous malformation 2 |
400 |
0.81 |
| chr10_11209293_11209598 | 1.13 |
CELF2 |
CUGBP, Elav-like family member 2 |
1954 |
0.35 |
| chr22_37542366_37542517 | 1.13 |
IL2RB |
interleukin 2 receptor, beta |
3589 |
0.14 |
| chr10_135202564_135202912 | 1.08 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
1600 |
0.18 |
| chr20_62368950_62369101 | 1.05 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
388 |
0.53 |
| chr13_52398191_52398536 | 1.02 |
RP11-327P2.5 |
|
19930 |
0.17 |
| chr19_40447118_40447269 | 1.01 |
FCGBP |
Fc fragment of IgG binding protein |
6660 |
0.17 |
| chr12_56415966_56416141 | 0.97 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
953 |
0.34 |
| chr16_85932806_85932957 | 0.97 |
IRF8 |
interferon regulatory factor 8 |
112 |
0.97 |
| chr11_6767215_6767876 | 0.97 |
OR2AG2 |
olfactory receptor, family 2, subfamily AG, member 2 |
22741 |
0.11 |
| chr14_92332597_92332814 | 0.94 |
TC2N |
tandem C2 domains, nuclear |
1168 |
0.51 |
| chr1_100887352_100887656 | 0.94 |
ENSG00000216067 |
. |
43173 |
0.14 |
| chr1_156458657_156458808 | 0.93 |
MEF2D |
myocyte enhancer factor 2D |
1659 |
0.26 |
| chr10_63663177_63663552 | 0.92 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
2305 |
0.38 |
| chr22_26823969_26824219 | 0.92 |
ASPHD2 |
aspartate beta-hydroxylase domain containing 2 |
1145 |
0.46 |
| chr6_32407696_32407883 | 0.87 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
134 |
0.95 |
| chr11_60850090_60850241 | 0.86 |
CD5 |
CD5 molecule |
19702 |
0.16 |
| chrY_16636064_16636659 | 0.86 |
NLGN4Y |
neuroligin 4, Y-linked |
93 |
0.99 |
| chr2_175462692_175462904 | 0.85 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
136 |
0.96 |
| chr2_43448588_43448818 | 0.84 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
5045 |
0.26 |
| chr1_150737696_150737976 | 0.83 |
CTSS |
cathepsin S |
432 |
0.77 |
| chr21_32557466_32557617 | 0.82 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
55002 |
0.16 |
| chr16_50303381_50303532 | 0.82 |
ADCY7 |
adenylate cyclase 7 |
2994 |
0.28 |
| chr12_15699329_15699635 | 0.79 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
196 |
0.97 |
| chr21_36715747_36715917 | 0.79 |
RUNX1 |
runt-related transcription factor 1 |
294191 |
0.01 |
| chr1_92951290_92951613 | 0.76 |
GFI1 |
growth factor independent 1 transcription repressor |
177 |
0.97 |
| chr3_151917198_151917573 | 0.75 |
MBNL1 |
muscleblind-like splicing regulator 1 |
68444 |
0.12 |
| chr5_150636818_150636969 | 0.75 |
GM2A |
GM2 ganglioside activator |
2467 |
0.28 |
| chr2_148602569_148602812 | 0.75 |
ACVR2A |
activin A receptor, type IIA |
505 |
0.82 |
| chr11_118209630_118210055 | 0.74 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
1810 |
0.22 |
| chr8_8727283_8727434 | 0.73 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
23797 |
0.18 |
| chr13_97874593_97875664 | 0.70 |
MBNL2 |
muscleblind-like splicing regulator 2 |
519 |
0.87 |
| chr20_31099035_31099434 | 0.69 |
C20orf112 |
chromosome 20 open reading frame 112 |
24 |
0.98 |
| chr21_43186131_43186315 | 0.69 |
RIPK4 |
receptor-interacting serine-threonine kinase 4 |
105 |
0.97 |
| chr12_51717169_51717830 | 0.68 |
BIN2 |
bridging integrator 2 |
400 |
0.82 |
| chr11_128395074_128395489 | 0.67 |
RP11-1007G5.2 |
|
756 |
0.69 |
| chr2_106043022_106043173 | 0.67 |
FHL2 |
four and a half LIM domains 2 |
11855 |
0.23 |
| chr4_89742277_89742465 | 0.66 |
FAM13A |
family with sequence similarity 13, member A |
1981 |
0.43 |
| chr5_1108899_1109050 | 0.66 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
3176 |
0.24 |
| chr10_104210626_104210853 | 0.65 |
ENSG00000269609 |
. |
351 |
0.57 |
| chr13_46754139_46754322 | 0.65 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
2229 |
0.26 |
| chr17_36864246_36864397 | 0.65 |
CTB-58E17.3 |
|
1807 |
0.14 |
| chr3_124303143_124303518 | 0.64 |
KALRN |
kalirin, RhoGEF kinase |
176 |
0.97 |
| chrX_51137751_51137902 | 0.64 |
RP11-348F1.3 |
|
1488 |
0.45 |
| chr2_242800719_242801011 | 0.63 |
PDCD1 |
programmed cell death 1 |
195 |
0.9 |
| chr16_54026157_54026308 | 0.63 |
RP11-357N13.2 |
|
7649 |
0.19 |
| chrX_154417087_154417278 | 0.62 |
VBP1 |
von Hippel-Lindau binding protein 1 |
8102 |
0.19 |
| chr9_130736579_130736730 | 0.61 |
FAM102A |
family with sequence similarity 102, member A |
6138 |
0.12 |
| chr2_191751090_191751325 | 0.61 |
AC005540.3 |
|
4422 |
0.24 |
| chr19_46219682_46220065 | 0.61 |
FBXO46 |
F-box protein 46 |
1337 |
0.23 |
| chr15_25200210_25200938 | 0.61 |
SNURF |
SNRPN upstream reading frame protein |
413 |
0.41 |
| chr1_9031254_9031405 | 0.60 |
ENSG00000265141 |
. |
8604 |
0.17 |
| chr1_209942353_209942874 | 0.59 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
653 |
0.63 |
| chr9_140356804_140356997 | 0.59 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
3114 |
0.16 |
| chr6_111302519_111302780 | 0.59 |
RPF2 |
ribosome production factor 2 homolog (S. cerevisiae) |
569 |
0.68 |
| chr7_13097458_13097609 | 0.58 |
ENSG00000222974 |
. |
181224 |
0.03 |
| chr14_22963855_22964006 | 0.58 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
7759 |
0.1 |
| chr4_100738641_100738957 | 0.58 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
796 |
0.72 |
| chr5_133451742_133452000 | 0.57 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
555 |
0.8 |
| chr1_198609469_198609717 | 0.57 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
1301 |
0.54 |
| chr14_21736745_21736945 | 0.57 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
35 |
0.97 |
| chr1_10759853_10760004 | 0.56 |
CASZ1 |
castor zinc finger 1 |
54795 |
0.12 |
| chr1_60162288_60162439 | 0.56 |
ENSG00000266150 |
. |
36605 |
0.2 |
| chr7_30634096_30634338 | 0.55 |
GARS |
glycyl-tRNA synthetase |
80 |
0.61 |
| chrX_70917395_70917655 | 0.55 |
ENSG00000221684 |
. |
61780 |
0.11 |
| chr6_144732729_144732880 | 0.55 |
UTRN |
utrophin |
67567 |
0.13 |
| chr1_26872331_26872899 | 0.55 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
272 |
0.88 |
| chr13_28715875_28716136 | 0.55 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
2835 |
0.24 |
| chr4_156760399_156760658 | 0.55 |
TDO2 |
tryptophan 2,3-dioxygenase |
15386 |
0.21 |
| chr17_47287660_47288689 | 0.54 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
238 |
0.65 |
| chr1_154915200_154915451 | 0.54 |
PMVK |
phosphomevalonate kinase |
5858 |
0.09 |
| chrX_106362200_106362401 | 0.53 |
RBM41 |
RNA binding motif protein 41 |
243 |
0.94 |
| chr8_97533683_97533834 | 0.53 |
SDC2 |
syndecan 2 |
27522 |
0.23 |
| chr7_154719533_154719892 | 0.53 |
PAXIP1-AS2 |
PAXIP1 antisense RNA 2 |
515 |
0.78 |
| chr15_89009981_89010294 | 0.53 |
MRPL46 |
mitochondrial ribosomal protein L46 |
513 |
0.47 |
| chr17_19280691_19280938 | 0.53 |
MAPK7 |
mitogen-activated protein kinase 7 |
220 |
0.65 |
| chr7_155437664_155437928 | 0.52 |
RBM33 |
RNA binding motif protein 33 |
400 |
0.67 |
| chr7_64541108_64541627 | 0.52 |
ENSG00000207062 |
. |
10451 |
0.15 |
| chr13_99226843_99226994 | 0.51 |
STK24 |
serine/threonine kinase 24 |
2199 |
0.3 |
| chr11_116644068_116644415 | 0.51 |
AP006216.10 |
|
136 |
0.83 |
| chr19_16379423_16379574 | 0.51 |
CTD-2562J15.6 |
|
24888 |
0.13 |
| chr3_185652121_185652403 | 0.51 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
3539 |
0.25 |
| chr18_60984919_60985484 | 0.51 |
BCL2 |
B-cell CLL/lymphoma 2 |
844 |
0.57 |
| chrX_71003427_71003880 | 0.50 |
ENSG00000221684 |
. |
24348 |
0.23 |
| chr20_30262987_30263252 | 0.50 |
RP11-243J16.7 |
|
10900 |
0.13 |
| chr11_8929144_8929295 | 0.50 |
ST5 |
suppression of tumorigenicity 5 |
3279 |
0.14 |
| chr12_111867666_111868155 | 0.49 |
SH2B3 |
SH2B adaptor protein 3 |
4756 |
0.21 |
| chr4_103423579_103423896 | 0.49 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
682 |
0.61 |
| chrY_15017679_15018174 | 0.49 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
277 |
0.95 |
| chr11_65647291_65647461 | 0.48 |
CTSW |
cathepsin W |
47 |
0.93 |
| chr7_128379466_128379711 | 0.48 |
CALU |
calumenin |
130 |
0.94 |
| chrX_111125134_111125285 | 0.48 |
TRPC5OS |
TRPC5 opposite strand |
84 |
0.98 |
| chr12_1920714_1920865 | 0.48 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
97 |
0.97 |
| chr11_67204819_67205147 | 0.48 |
PTPRCAP |
protein tyrosine phosphatase, receptor type, C-associated protein |
555 |
0.47 |
| chr19_50879744_50879984 | 0.48 |
NR1H2 |
nuclear receptor subfamily 1, group H, member 2 |
134 |
0.91 |
| chr8_26162809_26162960 | 0.47 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
11885 |
0.26 |
| chr2_106407332_106407557 | 0.47 |
NCK2 |
NCK adaptor protein 2 |
25571 |
0.24 |
| chr1_8685789_8685940 | 0.47 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
1425 |
0.54 |
| chr13_50158821_50159403 | 0.47 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
602 |
0.77 |
| chr2_85076559_85076967 | 0.47 |
TRABD2A |
TraB domain containing 2A |
31443 |
0.16 |
| chr11_67186882_67187131 | 0.47 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
1646 |
0.15 |
| chr1_100715442_100715812 | 0.47 |
DBT |
dihydrolipoamide branched chain transacylase E2 |
237 |
0.9 |
| chr21_36418295_36418575 | 0.47 |
RUNX1 |
runt-related transcription factor 1 |
3027 |
0.41 |
| chr21_34142699_34142850 | 0.47 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
1205 |
0.34 |
| chr19_12847748_12847986 | 0.47 |
ASNA1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
86 |
0.91 |
| chr10_64134972_64135123 | 0.46 |
ZNF365 |
zinc finger protein 365 |
256 |
0.94 |
| chr19_12906733_12906884 | 0.46 |
CTD-2659N19.10 |
|
829 |
0.32 |
| chr1_50513511_50513662 | 0.46 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
100 |
0.97 |
| chr3_16926768_16927235 | 0.46 |
PLCL2 |
phospholipase C-like 2 |
549 |
0.84 |
| chr18_9404902_9405053 | 0.46 |
RALBP1 |
ralA binding protein 1 |
70030 |
0.08 |
| chr20_61554036_61554389 | 0.45 |
DIDO1 |
death inducer-obliterator 1 |
3609 |
0.18 |
| chr3_129034345_129034731 | 0.45 |
H1FX-AS1 |
H1FX antisense RNA 1 |
175 |
0.82 |
| chrX_70838155_70838405 | 0.45 |
CXCR3 |
chemokine (C-X-C motif) receptor 3 |
80 |
0.97 |
| chr1_168523425_168523576 | 0.45 |
XCL2 |
chemokine (C motif) ligand 2 |
10265 |
0.23 |
| chr6_150038866_150039094 | 0.45 |
LATS1 |
large tumor suppressor kinase 1 |
190 |
0.93 |
| chr9_20622216_20622410 | 0.45 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
201 |
0.95 |
| chr3_172232547_172232820 | 0.44 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
8582 |
0.25 |
| chr4_151995452_151995603 | 0.44 |
RPS3A |
ribosomal protein S3A |
25198 |
0.14 |
| chr10_5855917_5856253 | 0.44 |
GDI2 |
GDP dissociation inhibitor 2 |
207 |
0.94 |
| chr2_87214834_87214999 | 0.43 |
PLGLB1 |
plasminogen-like B1 |
34059 |
0.19 |
| chr4_6576981_6577196 | 0.43 |
MAN2B2 |
mannosidase, alpha, class 2B, member 2 |
146 |
0.96 |
| chr19_58898981_58899504 | 0.43 |
RPS5 |
ribosomal protein S5 |
586 |
0.41 |
| chr15_93457570_93457977 | 0.43 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
10070 |
0.2 |
| chr14_22972748_22972969 | 0.43 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
16687 |
0.09 |
| chr2_88081579_88081730 | 0.42 |
PLGLB2 |
plasminogen-like B2 |
34048 |
0.19 |
| chr11_17298295_17298504 | 0.42 |
NUCB2 |
nucleobindin 2 |
21 |
0.98 |
| chr17_60802287_60802509 | 0.42 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
11480 |
0.17 |
| chr13_113807206_113807543 | 0.42 |
PROZ |
protein Z, vitamin K-dependent plasma glycoprotein |
5594 |
0.13 |
| chr7_26192485_26193339 | 0.42 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
1052 |
0.53 |
| chr6_159420910_159421511 | 0.42 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
9 |
0.82 |
| chr7_99096274_99096688 | 0.42 |
ZNF394 |
zinc finger protein 394 |
1406 |
0.25 |
| chr12_55373639_55373790 | 0.42 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
1908 |
0.39 |
| chr11_111798023_111798431 | 0.42 |
DIXDC1 |
DIX domain containing 1 |
359 |
0.67 |
| chr16_2015079_2015314 | 0.42 |
ENSG00000255198 |
. |
11 |
0.78 |
| chr1_226068030_226068371 | 0.42 |
TMEM63A |
transmembrane protein 63A |
1869 |
0.24 |
| chr12_4938445_4938596 | 0.41 |
RP3-377H17.2 |
|
9601 |
0.2 |
| chr4_38666789_38667086 | 0.41 |
RP11-617D20.1 |
|
433 |
0.48 |
| chr2_110595491_110595642 | 0.41 |
RGPD5 |
RANBP2-like and GRIP domain containing 5 |
43304 |
0.17 |
| chr14_64107873_64108536 | 0.41 |
WDR89 |
WD repeat domain 89 |
79 |
0.96 |
| chr2_108489338_108489489 | 0.41 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
46020 |
0.19 |
| chr19_48835680_48835937 | 0.41 |
EMP3 |
epithelial membrane protein 3 |
6943 |
0.11 |
| chr9_80523811_80523962 | 0.41 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
85971 |
0.1 |
| chr17_38559088_38559385 | 0.41 |
ENSG00000222881 |
. |
11646 |
0.11 |
| chr11_124618587_124618738 | 0.41 |
RP11-677M14.2 |
|
2429 |
0.15 |
| chr19_50651113_50651549 | 0.41 |
IZUMO2 |
IZUMO family member 2 |
15082 |
0.15 |
| chr6_11382632_11382872 | 0.40 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
171 |
0.97 |
| chr3_39321282_39321554 | 0.40 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
107 |
0.97 |
| chr12_14518062_14518320 | 0.40 |
ATF7IP |
activating transcription factor 7 interacting protein |
425 |
0.88 |
| chr8_22463688_22463839 | 0.40 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
350 |
0.76 |
| chr10_106027925_106028215 | 0.40 |
ENSG00000266852 |
. |
107 |
0.87 |
| chr2_169458843_169458994 | 0.40 |
ENSG00000199348 |
. |
4088 |
0.27 |
| chr2_113145556_113145707 | 0.40 |
RGPD8 |
RANBP2-like and GRIP domain containing 8 |
45465 |
0.15 |
| chr1_198637715_198637866 | 0.39 |
RP11-553K8.5 |
|
1600 |
0.47 |
| chr12_54811406_54811839 | 0.39 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
1622 |
0.22 |
| chr6_26442341_26442492 | 0.39 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
1630 |
0.25 |
| chr15_68570739_68571112 | 0.39 |
FEM1B |
fem-1 homolog b (C. elegans) |
784 |
0.64 |
| chr1_43803771_43803922 | 0.39 |
MPL |
myeloproliferative leukemia virus oncogene |
326 |
0.8 |
| chr1_21859764_21859915 | 0.39 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
17933 |
0.19 |
| chr6_29692032_29692183 | 0.39 |
HLA-F |
major histocompatibility complex, class I, F |
291 |
0.87 |
| chr2_111291029_111291180 | 0.39 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
483 |
0.83 |
| chr9_127950035_127950361 | 0.39 |
PPP6C |
protein phosphatase 6, catalytic subunit |
1864 |
0.3 |
| chr16_80838686_80839030 | 0.39 |
CDYL2 |
chromodomain protein, Y-like 2 |
632 |
0.79 |
| chr21_30397725_30398151 | 0.39 |
RP1-100J12.1 |
|
727 |
0.42 |
| chr5_134181747_134182027 | 0.38 |
C5orf24 |
chromosome 5 open reading frame 24 |
43 |
0.97 |
| chr20_13764930_13765215 | 0.38 |
ESF1 |
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
460 |
0.52 |
| chr5_154137578_154138162 | 0.38 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
1136 |
0.49 |
| chr1_152162233_152162399 | 0.38 |
FLG-AS1 |
FLG antisense RNA 1 |
489 |
0.77 |
| chr6_134861028_134861179 | 0.38 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
221853 |
0.02 |
| chr2_28616306_28616795 | 0.37 |
FOSL2 |
FOS-like antigen 2 |
825 |
0.43 |
| chr22_50523508_50523892 | 0.37 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
12 |
0.97 |
| chr11_559952_560103 | 0.37 |
RASSF7 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
377 |
0.43 |
| chr3_141121878_141122254 | 0.37 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
406 |
0.88 |
| chr2_106367063_106367214 | 0.37 |
NCK2 |
NCK adaptor protein 2 |
4950 |
0.33 |
| chr3_160167041_160167544 | 0.37 |
TRIM59 |
tripartite motif containing 59 |
109 |
0.96 |
| chr16_8715324_8715502 | 0.37 |
METTL22 |
methyltransferase like 22 |
127 |
0.97 |
| chr19_47125438_47125589 | 0.37 |
PTGIR |
prostaglandin I2 (prostacyclin) receptor (IP) |
2781 |
0.13 |
| chr10_43902220_43902565 | 0.37 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
875 |
0.54 |
| chr9_134405336_134405487 | 0.37 |
UCK1 |
uridine-cytidine kinase 1 |
1155 |
0.4 |
| chr11_67176589_67176876 | 0.37 |
TBC1D10C |
TBC1 domain family, member 10C |
5072 |
0.07 |
| chr2_231191950_231192650 | 0.36 |
SP140L |
SP140 nuclear body protein-like |
315 |
0.92 |
| chr1_170536747_170536898 | 0.36 |
RP11-576I22.2 |
|
35034 |
0.18 |
| chr16_3660969_3661192 | 0.36 |
SLX4 |
SLX4 structure-specific endonuclease subunit |
519 |
0.73 |
| chr19_10675577_10675931 | 0.36 |
KRI1 |
KRI1 homolog (S. cerevisiae) |
912 |
0.35 |
| chr1_212588032_212588228 | 0.35 |
TMEM206 |
transmembrane protein 206 |
107 |
0.96 |
| chr2_118572851_118573368 | 0.35 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
883 |
0.63 |
| chr2_98261858_98262243 | 0.35 |
COX5B |
cytochrome c oxidase subunit Vb |
453 |
0.77 |
| chr9_130692852_130693086 | 0.35 |
PIP5KL1 |
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
107 |
0.92 |
| chr2_204735521_204735672 | 0.35 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
641 |
0.79 |
| chr3_71630406_71630763 | 0.35 |
FOXP1 |
forkhead box P1 |
2320 |
0.31 |
| chr1_59164702_59165010 | 0.35 |
MYSM1 |
Myb-like, SWIRM and MPN domains 1 |
908 |
0.62 |
| chr12_66524690_66525029 | 0.35 |
LLPH |
LLP homolog, long-term synaptic facilitation (Aplysia) |
311 |
0.49 |
| chr1_32721190_32721341 | 0.35 |
LCK |
lymphocyte-specific protein tyrosine kinase |
4390 |
0.1 |
| chr9_130536234_130536385 | 0.35 |
SH2D3C |
SH2 domain containing 3C |
2592 |
0.13 |
| chr4_4290910_4291202 | 0.35 |
LYAR |
Ly1 antibody reactive |
692 |
0.48 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.4 | 1.3 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.4 | 1.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.2 | 0.7 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.2 | 0.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.2 | 0.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.4 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.3 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.3 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.1 | 0.5 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.1 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.1 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.1 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.1 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 1.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.2 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.4 | GO:1904377 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.3 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.6 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.2 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.5 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 1.0 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 | 0.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.4 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.5 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.2 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.3 | GO:1902579 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0010823 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.7 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.0 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.0 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 1.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.2 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 0.5 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:1903578 | regulation of glycolytic process(GO:0006110) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.5 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 2.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:1990391 | mismatch repair complex(GO:0032300) DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 1.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.1 | 0.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.3 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.3 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 1.2 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.4 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 0.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |