Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
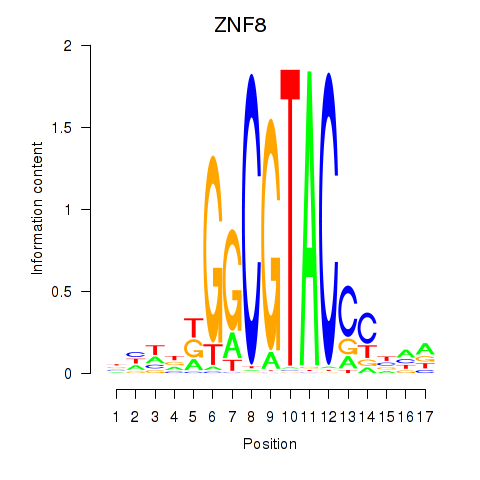
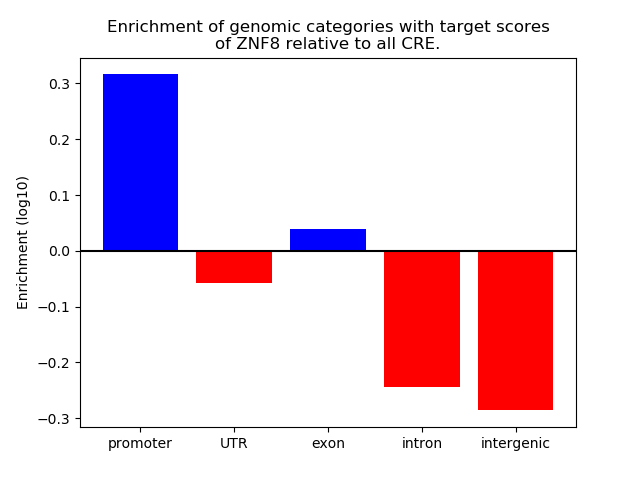
Results for ZNF8
Z-value: 1.02
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_58791267_58791418 | ZNF8 | 1024 | 0.277306 | 0.82 | 7.3e-03 | Click! |
| chr19_58790338_58791095 | ZNF8 | 398 | 0.513430 | 0.59 | 9.5e-02 | Click! |
| chr19_58805596_58805747 | ZNF8 | 15353 | 0.078411 | -0.05 | 9.1e-01 | Click! |
Activity of the ZNF8 motif across conditions
Conditions sorted by the z-value of the ZNF8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
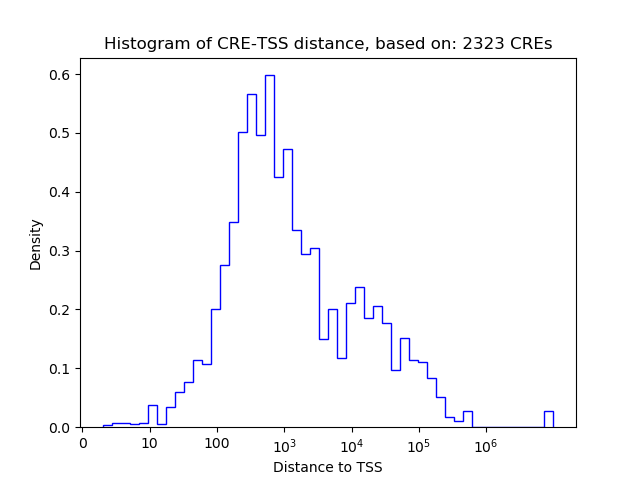
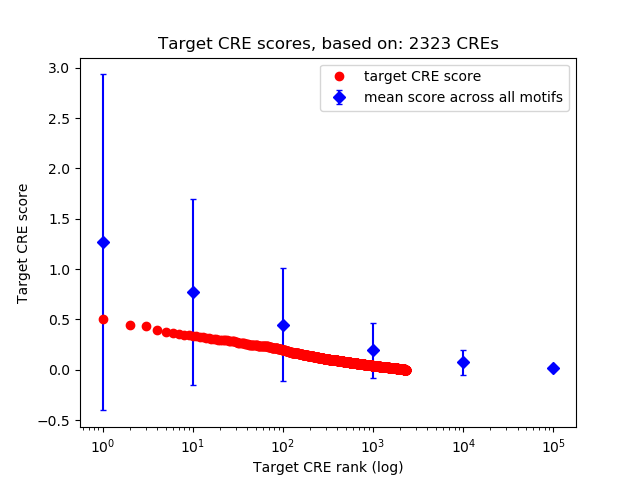
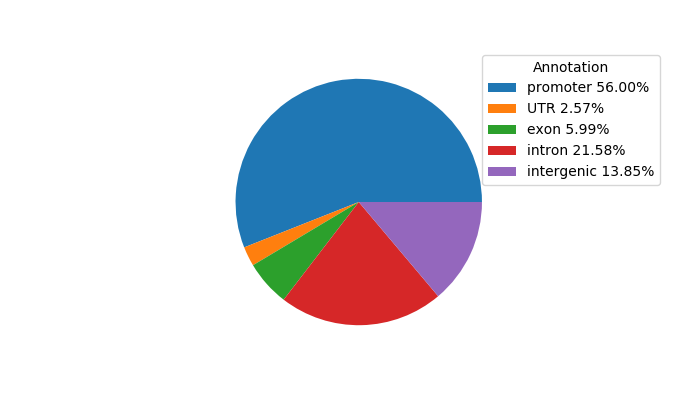
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_12575381_12575532 | 0.51 |
TYRP1 |
tyrosinase-related protein 1 |
109983 |
0.07 |
| chr17_7607729_7607918 | 0.45 |
EFNB3 |
ephrin-B3 |
697 |
0.47 |
| chr17_57439920_57440434 | 0.44 |
ENSG00000263857 |
. |
3267 |
0.22 |
| chr10_135193239_135193390 | 0.39 |
AL360181.1 |
Uncharacterized protein |
212 |
0.71 |
| chr7_72974672_72974823 | 0.38 |
BCL7B |
B-cell CLL/lymphoma 7B |
2415 |
0.22 |
| chr6_119029777_119030724 | 0.37 |
CEP85L |
centrosomal protein 85kDa-like |
981 |
0.69 |
| chr12_104851638_104851843 | 0.36 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
961 |
0.68 |
| chr17_47271115_47271274 | 0.35 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
15080 |
0.14 |
| chr3_114014633_114014828 | 0.35 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
896 |
0.58 |
| chr9_95055581_95055984 | 0.34 |
IARS |
isoleucyl-tRNA synthetase |
141 |
0.91 |
| chr1_36042129_36042429 | 0.34 |
RP4-728D4.2 |
|
1051 |
0.43 |
| chr1_172109378_172109611 | 0.33 |
ENSG00000207949 |
. |
1447 |
0.39 |
| chr8_117884616_117884915 | 0.33 |
RAD21 |
RAD21 homolog (S. pombe) |
1834 |
0.24 |
| chr21_17681930_17682081 | 0.32 |
ENSG00000201025 |
. |
24916 |
0.27 |
| chr14_23398675_23398826 | 0.32 |
PRMT5 |
protein arginine methyltransferase 5 |
44 |
0.51 |
| chr11_105392577_105392728 | 0.31 |
GRIA4 |
glutamate receptor, ionotropic, AMPA 4 |
88069 |
0.1 |
| chr15_89179199_89179367 | 0.30 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
101 |
0.96 |
| chrX_72783288_72783711 | 0.30 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
455 |
0.89 |
| chr2_235403849_235404784 | 0.30 |
ARL4C |
ADP-ribosylation factor-like 4C |
928 |
0.74 |
| chr17_71088602_71088815 | 0.30 |
SLC39A11 |
solute carrier family 39, member 11 |
107 |
0.98 |
| chr6_28180364_28180829 | 0.30 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
12068 |
0.13 |
| chrX_122318159_122318585 | 0.30 |
GRIA3 |
glutamate receptor, ionotropic, AMPA 3 |
36 |
0.99 |
| chr1_19281880_19282035 | 0.29 |
IFFO2 |
intermediate filament family orphan 2 |
235 |
0.94 |
| chr19_41732107_41732593 | 0.29 |
AXL |
AXL receptor tyrosine kinase |
310 |
0.83 |
| chr4_81124101_81124367 | 0.29 |
PRDM8 |
PR domain containing 8 |
5576 |
0.24 |
| chr10_116584415_116584656 | 0.29 |
FAM160B1 |
family with sequence similarity 160, member B1 |
3006 |
0.37 |
| chr20_1099338_1099605 | 0.29 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
191 |
0.95 |
| chr13_19203049_19203245 | 0.29 |
ENSG00000223024 |
. |
240284 |
0.02 |
| chr5_43192499_43192793 | 0.28 |
NIM1K |
NIM1 serine/threonine protein kinase |
308 |
0.92 |
| chr7_43945567_43945756 | 0.28 |
URGCP-MRPS24 |
URGCP-MRPS24 readthrough |
527 |
0.44 |
| chr21_26934419_26935339 | 0.28 |
ENSG00000234883 |
. |
11413 |
0.19 |
| chr16_66652344_66652579 | 0.27 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
13037 |
0.11 |
| chr5_134210294_134210605 | 0.27 |
TXNDC15 |
thioredoxin domain containing 15 |
337 |
0.87 |
| chr12_26844097_26844392 | 0.27 |
RP11-666F17.1 |
|
67942 |
0.12 |
| chr17_27169901_27170114 | 0.26 |
FAM222B |
family with sequence similarity 222, member B |
150 |
0.9 |
| chrX_106243219_106243547 | 0.26 |
MORC4 |
MORC family CW-type zinc finger 4 |
75 |
0.98 |
| chr19_17357023_17357252 | 0.26 |
NR2F6 |
nuclear receptor subfamily 2, group F, member 6 |
388 |
0.67 |
| chr6_158589460_158589825 | 0.26 |
GTF2H5 |
general transcription factor IIH, polypeptide 5 |
258 |
0.57 |
| chr10_72201220_72201371 | 0.26 |
NODAL |
nodal growth differentiation factor |
128 |
0.96 |
| chr12_21606326_21606605 | 0.26 |
PYROXD1 |
pyridine nucleotide-disulphide oxidoreductase domain 1 |
15616 |
0.17 |
| chr11_14520450_14521043 | 0.25 |
COPB1 |
coatomer protein complex, subunit beta 1 |
611 |
0.75 |
| chr10_105605562_105605713 | 0.25 |
SH3PXD2A |
SH3 and PX domains 2A |
9527 |
0.2 |
| chr19_34012835_34013082 | 0.25 |
PEPD |
peptidase D |
258 |
0.95 |
| chr19_37407398_37407734 | 0.25 |
ZNF568 |
zinc finger protein 568 |
209 |
0.67 |
| chr10_94334737_94334888 | 0.25 |
IDE |
insulin-degrading enzyme |
979 |
0.59 |
| chr16_49889200_49889463 | 0.25 |
ZNF423 |
zinc finger protein 423 |
715 |
0.75 |
| chr2_197124773_197125094 | 0.25 |
AC020571.3 |
|
95 |
0.97 |
| chr12_120876193_120876532 | 0.25 |
AL021546.6 |
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
361 |
0.52 |
| chr16_4818494_4818716 | 0.25 |
ZNF500 |
zinc finger protein 500 |
980 |
0.39 |
| chr14_33929813_33929964 | 0.25 |
NPAS3 |
neuronal PAS domain protein 3 |
245370 |
0.02 |
| chr11_118964238_118964465 | 0.24 |
H2AFX |
H2A histone family, member X |
1826 |
0.14 |
| chr3_52737763_52738047 | 0.24 |
GLT8D1 |
glycosyltransferase 8 domain containing 1 |
270 |
0.75 |
| chr17_78711366_78711517 | 0.24 |
RP11-28G8.1 |
|
67991 |
0.11 |
| chr19_39835748_39835899 | 0.24 |
SAMD4B |
sterile alpha motif domain containing 4B |
2715 |
0.12 |
| chr5_127420764_127420915 | 0.24 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
1299 |
0.63 |
| chr13_99958437_99958588 | 0.24 |
GPR183 |
G protein-coupled receptor 183 |
1147 |
0.52 |
| chr2_30458178_30458329 | 0.24 |
LBH |
limb bud and heart development |
3207 |
0.3 |
| chr9_123555070_123555612 | 0.24 |
FBXW2 |
F-box and WD repeat domain containing 2 |
314 |
0.9 |
| chr5_110559444_110559831 | 0.24 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
11 |
0.98 |
| chrX_53251626_53252025 | 0.24 |
KDM5C |
lysine (K)-specific demethylase 5C |
2533 |
0.26 |
| chr3_197477598_197478224 | 0.23 |
FYTTD1 |
forty-two-three domain containing 1 |
650 |
0.6 |
| chr17_3598807_3599359 | 0.23 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
244 |
0.86 |
| chr20_57427056_57427350 | 0.23 |
GNAS |
GNAS complex locus |
566 |
0.59 |
| chr19_45251774_45251925 | 0.23 |
BCL3 |
B-cell CLL/lymphoma 3 |
45 |
0.96 |
| chr12_6184333_6184641 | 0.23 |
ENSG00000240533 |
. |
20649 |
0.2 |
| chr10_105880540_105880741 | 0.23 |
SFR1 |
SWI5-dependent recombination repair 1 |
1176 |
0.46 |
| chr18_14131976_14132486 | 0.23 |
ZNF519 |
zinc finger protein 519 |
196 |
0.93 |
| chr12_55375935_55376187 | 0.23 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
439 |
0.86 |
| chr2_65577328_65577479 | 0.23 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
5483 |
0.26 |
| chr10_5727764_5728131 | 0.23 |
FAM208B |
family with sequence similarity 208, member B |
1036 |
0.5 |
| chr13_25861194_25861587 | 0.23 |
MTMR6 |
myotubularin related protein 6 |
314 |
0.89 |
| chr16_66461764_66462073 | 0.23 |
BEAN1 |
brain expressed, associated with NEDD4, 1 |
644 |
0.68 |
| chr15_85523909_85524155 | 0.23 |
PDE8A |
phosphodiesterase 8A |
288 |
0.91 |
| chr12_52463075_52463333 | 0.22 |
C12orf44 |
chromosome 12 open reading frame 44 |
551 |
0.63 |
| chr15_25201119_25201420 | 0.22 |
SNURF |
SNRPN upstream reading frame protein |
1108 |
0.23 |
| chr1_151811278_151811720 | 0.22 |
C2CD4D |
C2 calcium-dependent domain containing 4D |
1534 |
0.18 |
| chr5_138676935_138677379 | 0.22 |
PAIP2 |
poly(A) binding protein interacting protein 2 |
119 |
0.93 |
| chr14_90865510_90865807 | 0.22 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
1052 |
0.5 |
| chrX_92928498_92928999 | 0.22 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
181 |
0.61 |
| chr2_96809635_96809786 | 0.21 |
DUSP2 |
dual specificity phosphatase 2 |
1469 |
0.34 |
| chr1_70670699_70671189 | 0.21 |
LRRC40 |
leucine rich repeat containing 40 |
359 |
0.53 |
| chr1_236960314_236960595 | 0.21 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
1472 |
0.52 |
| chr8_38648804_38648992 | 0.21 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
3217 |
0.23 |
| chr16_72128402_72128590 | 0.21 |
TXNL4B |
thioredoxin-like 4B |
166 |
0.84 |
| chr14_61811311_61811517 | 0.21 |
PRKCH |
protein kinase C, eta |
372 |
0.88 |
| chr8_124172522_124172673 | 0.21 |
FAM83A |
family with sequence similarity 83, member A |
18690 |
0.11 |
| chr5_10353857_10354094 | 0.21 |
MARCH6 |
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
96 |
0.72 |
| chr15_68545515_68545666 | 0.21 |
CLN6 |
ceroid-lipofuscinosis, neuronal 6, late infantile, variant |
3854 |
0.2 |
| chr12_125424789_125424964 | 0.21 |
UBC |
ubiquitin C |
22962 |
0.14 |
| chr1_40420311_40420468 | 0.21 |
MFSD2A |
major facilitator superfamily domain containing 2A |
413 |
0.81 |
| chr10_497094_497245 | 0.21 |
RP11-490E15.2 |
|
13668 |
0.2 |
| chr3_104938786_104938937 | 0.21 |
ALCAM |
activated leukocyte cell adhesion molecule |
146892 |
0.05 |
| chr2_73612447_73612786 | 0.21 |
ALMS1 |
Alstrom syndrome 1 |
270 |
0.93 |
| chr12_92949870_92950021 | 0.20 |
ENSG00000238865 |
. |
11182 |
0.27 |
| chrX_130946561_130947407 | 0.20 |
ENSG00000200587 |
. |
126000 |
0.06 |
| chr6_104730379_104730530 | 0.20 |
ENSG00000252944 |
. |
484243 |
0.01 |
| chr2_194347562_194347833 | 0.20 |
NA |
NA |
> 106 |
NA |
| chr10_95653765_95654590 | 0.20 |
SLC35G1 |
solute carrier family 35, member G1 |
447 |
0.88 |
| chr9_34178665_34178872 | 0.20 |
UBAP1 |
ubiquitin associated protein 1 |
235 |
0.91 |
| chr1_228593151_228593670 | 0.20 |
TRIM11 |
tripartite motif containing 11 |
195 |
0.86 |
| chr5_176023650_176023811 | 0.20 |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
13404 |
0.11 |
| chrY_17567786_17567937 | 0.20 |
ENSG00000252664 |
. |
606785 |
0.0 |
| chr9_98279774_98279925 | 0.20 |
PTCH1 |
patched 1 |
510 |
0.75 |
| chr12_48745022_48745253 | 0.20 |
ZNF641 |
zinc finger protein 641 |
60 |
0.85 |
| chr20_34681844_34681995 | 0.19 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
1160 |
0.49 |
| chr22_25506661_25506812 | 0.19 |
CTA-221G9.11 |
|
1923 |
0.39 |
| chr17_46633195_46633347 | 0.19 |
HOXB3 |
homeobox B3 |
612 |
0.44 |
| chr12_40012503_40012670 | 0.19 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
967 |
0.63 |
| chr2_133402716_133402880 | 0.19 |
AC010974.3 |
|
15301 |
0.22 |
| chr1_246670350_246670537 | 0.19 |
SMYD3 |
SET and MYND domain containing 3 |
76 |
0.98 |
| chr21_16427122_16427273 | 0.19 |
NRIP1 |
nuclear receptor interacting protein 1 |
9929 |
0.27 |
| chr2_142888569_142888850 | 0.19 |
LRP1B |
low density lipoprotein receptor-related protein 1B |
135 |
0.97 |
| chr12_90100416_90100567 | 0.19 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
2117 |
0.33 |
| chr12_56881156_56881307 | 0.18 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
671 |
0.58 |
| chr1_243584631_243584835 | 0.18 |
ENSG00000265201 |
. |
75255 |
0.11 |
| chr14_22409532_22409683 | 0.18 |
ENSG00000222776 |
. |
160822 |
0.03 |
| chr5_175621560_175621795 | 0.18 |
SIMC1 |
SUMO-interacting motifs containing 1 |
43688 |
0.14 |
| chr1_27126960_27127111 | 0.18 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
11616 |
0.11 |
| chr19_36035577_36035728 | 0.18 |
AD000090.2 |
|
290 |
0.72 |
| chr13_107220642_107220857 | 0.18 |
ARGLU1 |
arginine and glutamate rich 1 |
237 |
0.95 |
| chr11_62622399_62622788 | 0.18 |
ENSG00000255717 |
. |
38 |
0.91 |
| chr19_45312390_45312692 | 0.18 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
163 |
0.92 |
| chr18_67614161_67614403 | 0.17 |
CD226 |
CD226 molecule |
373 |
0.92 |
| chr10_97667413_97667615 | 0.17 |
ENTPD1-AS1 |
ENTPD1 antisense RNA 1 |
18 |
0.66 |
| chr6_26158769_26159181 | 0.17 |
HIST1H2BD |
histone cluster 1, H2bd |
601 |
0.45 |
| chr12_122326288_122326449 | 0.17 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
269 |
0.88 |
| chr1_11845061_11845371 | 0.17 |
C1orf167 |
chromosome 1 open reading frame 167 |
5342 |
0.12 |
| chr6_24743062_24743323 | 0.17 |
C6orf62 |
chromosome 6 open reading frame 62 |
22128 |
0.12 |
| chr11_28119423_28119574 | 0.17 |
METTL15 |
methyltransferase like 15 |
10297 |
0.21 |
| chrX_1653880_1654070 | 0.17 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
2025 |
0.36 |
| chr12_27090977_27091227 | 0.17 |
ASUN |
asunder spermatogenesis regulator |
135 |
0.61 |
| chr11_48041704_48041855 | 0.17 |
AC103828.1 |
|
4372 |
0.25 |
| chr5_49736865_49737170 | 0.17 |
EMB |
embigin |
167 |
0.98 |
| chr9_99181784_99182155 | 0.17 |
ZNF367 |
zinc finger protein 367 |
1358 |
0.46 |
| chr8_61429545_61429985 | 0.17 |
RAB2A |
RAB2A, member RAS oncogene family |
25 |
0.98 |
| chrY_1603880_1604065 | 0.17 |
NA |
NA |
> 106 |
NA |
| chr1_72751044_72751264 | 0.17 |
NEGR1 |
neuronal growth regulator 1 |
2737 |
0.44 |
| chr19_4969639_4969870 | 0.16 |
KDM4B |
lysine (K)-specific demethylase 4B |
611 |
0.72 |
| chr6_46293041_46293192 | 0.16 |
RCAN2 |
regulator of calcineurin 2 |
289 |
0.94 |
| chr2_225448726_225449293 | 0.16 |
CUL3 |
cullin 3 |
1056 |
0.69 |
| chr12_50135384_50135571 | 0.16 |
TMBIM6 |
transmembrane BAX inhibitor motif containing 6 |
86 |
0.97 |
| chr15_64915060_64915211 | 0.16 |
ENSG00000207223 |
. |
29952 |
0.13 |
| chr6_30684473_30685093 | 0.16 |
MDC1 |
mediator of DNA-damage checkpoint 1 |
115 |
0.89 |
| chr2_149403004_149403363 | 0.16 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
194 |
0.97 |
| chr8_13123268_13123419 | 0.16 |
DLC1 |
deleted in liver cancer 1 |
10712 |
0.27 |
| chr21_35287147_35287573 | 0.16 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
478 |
0.61 |
| chr21_46289667_46289837 | 0.16 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
3854 |
0.16 |
| chr2_9697178_9697329 | 0.16 |
ADAM17 |
ADAM metallopeptidase domain 17 |
1332 |
0.42 |
| chr17_34136054_34136260 | 0.16 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
302 |
0.81 |
| chr7_137685587_137685789 | 0.16 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
1105 |
0.53 |
| chr7_48018318_48018852 | 0.16 |
HUS1 |
HUS1 checkpoint homolog (S. pombe) |
516 |
0.79 |
| chrX_131341527_131341678 | 0.16 |
RAP2C-AS1 |
RAP2C antisense RNA 1 |
9573 |
0.22 |
| chr7_157131771_157131922 | 0.16 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
669 |
0.77 |
| chr1_49242211_49242511 | 0.16 |
BEND5 |
BEN domain containing 5 |
229 |
0.96 |
| chr16_30640046_30640198 | 0.16 |
ZNF689 |
zinc finger protein 689 |
18422 |
0.07 |
| chr4_6711000_6711263 | 0.16 |
MRFAP1L1 |
Morf4 family associated protein 1-like 1 |
476 |
0.75 |
| chr19_58459192_58459462 | 0.16 |
ZNF256 |
zinc finger protein 256 |
284 |
0.84 |
| chr17_61523601_61523752 | 0.16 |
CYB561 |
cytochrome b561 |
9 |
0.97 |
| chr6_160560270_160560421 | 0.16 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
9552 |
0.25 |
| chr5_95992763_95992914 | 0.16 |
CAST |
calpastatin |
4939 |
0.22 |
| chr16_57496565_57496748 | 0.15 |
POLR2C |
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
357 |
0.81 |
| chr22_45664670_45664855 | 0.15 |
UPK3A |
uroplakin 3A |
16101 |
0.16 |
| chr13_20701746_20702011 | 0.15 |
GJA3 |
gap junction protein, alpha 3, 46kDa |
33310 |
0.16 |
| chr16_21964348_21964572 | 0.15 |
UQCRC2 |
ubiquinol-cytochrome c reductase core protein II |
232 |
0.92 |
| chr6_26056150_26056643 | 0.15 |
HIST1H1C |
histone cluster 1, H1c |
303 |
0.74 |
| chr11_118973164_118973374 | 0.15 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
417 |
0.63 |
| chr2_129075768_129075960 | 0.15 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
287 |
0.93 |
| chr19_37063422_37063596 | 0.15 |
AC092295.7 |
|
463 |
0.54 |
| chr6_90067633_90067883 | 0.15 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
5191 |
0.22 |
| chr17_9480443_9480594 | 0.15 |
WDR16 |
WD repeat domain 16 |
519 |
0.48 |
| chr19_17941346_17941497 | 0.15 |
INSL3 |
insulin-like 3 (Leydig cell) |
9038 |
0.11 |
| chr16_74641159_74641577 | 0.15 |
GLG1 |
golgi glycoprotein 1 |
356 |
0.9 |
| chr6_79788386_79788977 | 0.15 |
PHIP |
pleckstrin homology domain interacting protein |
728 |
0.79 |
| chr6_26032878_26033311 | 0.15 |
HIST1H2AB |
histone cluster 1, H2ab |
702 |
0.31 |
| chr2_172289877_172290488 | 0.15 |
METTL8 |
methyltransferase like 8 |
318 |
0.7 |
| chr7_112429598_112430043 | 0.15 |
TMEM168 |
transmembrane protein 168 |
614 |
0.84 |
| chr1_41177230_41177555 | 0.15 |
NFYC |
nuclear transcription factor Y, gamma |
2177 |
0.25 |
| chr3_47820837_47820988 | 0.15 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
2495 |
0.25 |
| chr17_45899176_45899438 | 0.15 |
OSBPL7 |
oxysterol binding protein-like 7 |
107 |
0.94 |
| chr20_47803254_47804087 | 0.15 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
1234 |
0.47 |
| chr9_134282757_134283214 | 0.15 |
PRRC2B |
proline-rich coiled-coil 2B |
13505 |
0.2 |
| chr21_15096270_15096556 | 0.15 |
AL050303.1 |
Uncharacterized protein; cDNA FLJ43689 fis, clone TBAES2003550 |
42954 |
0.16 |
| chr10_103814200_103814517 | 0.15 |
C10orf76 |
chromosome 10 open reading frame 76 |
1592 |
0.34 |
| chr7_39989218_39989369 | 0.15 |
RP11-467D6.1 |
|
61 |
0.83 |
| chr4_89204547_89205147 | 0.14 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
195 |
0.93 |
| chr6_164530063_164530741 | 0.14 |
ENSG00000266128 |
. |
268809 |
0.02 |
| chr5_67510861_67511301 | 0.14 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
467 |
0.86 |
| chr2_113341475_113341929 | 0.14 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
316 |
0.87 |
| chr12_125050945_125051262 | 0.14 |
NCOR2 |
nuclear receptor corepressor 2 |
907 |
0.73 |
| chr5_172197544_172197695 | 0.14 |
DUSP1 |
dual specificity phosphatase 1 |
579 |
0.67 |
| chr15_29131227_29131499 | 0.14 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
54 |
0.98 |
| chr6_82462588_82463015 | 0.14 |
FAM46A |
family with sequence similarity 46, member A |
310 |
0.9 |
| chr6_137365071_137365326 | 0.14 |
IL20RA |
interleukin 20 receptor, alpha |
213 |
0.95 |
| chr2_68946943_68947094 | 0.14 |
ARHGAP25 |
Rho GTPase activating protein 25 |
9384 |
0.26 |
| chr14_61790070_61790226 | 0.14 |
ENSG00000247287 |
. |
612 |
0.51 |
| chr17_70715496_70715723 | 0.14 |
ENSG00000222545 |
. |
4999 |
0.28 |
| chr11_66081550_66081990 | 0.14 |
RP11-867G23.13 |
|
1446 |
0.16 |
| chr6_157469997_157470201 | 0.14 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
54 |
0.98 |
| chr4_86702026_86702177 | 0.14 |
ARHGAP24 |
Rho GTPase activating protein 24 |
2242 |
0.41 |
| chr14_24584036_24584230 | 0.14 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
63 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0002834 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0010664 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0033233 | regulation of protein sumoylation(GO:0033233) |
| 0.0 | 0.3 | GO:0007215 | glutamate receptor signaling pathway(GO:0007215) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.0 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
| 0.0 | 0.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.0 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0001614 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |