Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
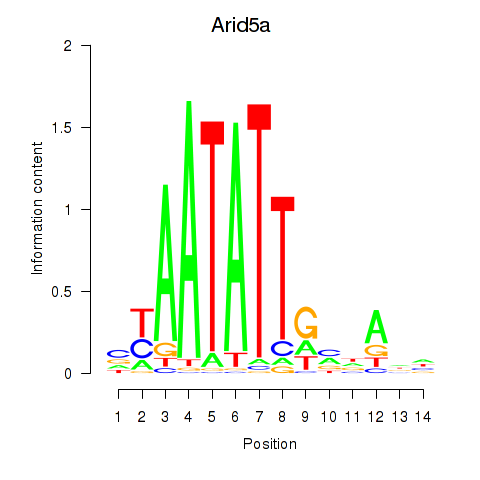
Results for Arid5a
Z-value: 0.40
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSMUSG00000037447.10 | AT rich interactive domain 5A (MRF1-like) |
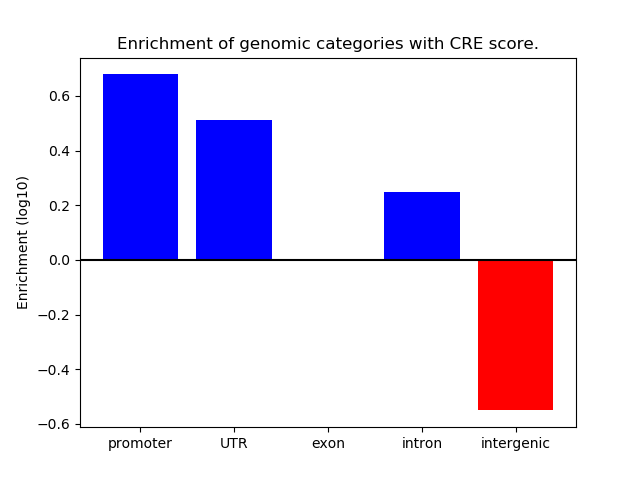
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_36307790_36308834 | Arid5a | 533 | 0.701103 | 0.21 | 1.0e-01 | Click! |
| chr1_36303019_36304082 | Arid5a | 4183 | 0.160223 | 0.19 | 1.4e-01 | Click! |
| chr1_36309796_36309947 | Arid5a | 2092 | 0.231507 | -0.10 | 4.7e-01 | Click! |
| chr1_36296960_36297111 | Arid5a | 10698 | 0.134106 | -0.09 | 5.0e-01 | Click! |
| chr1_36309057_36309739 | Arid5a | 1619 | 0.287283 | 0.00 | 9.8e-01 | Click! |
Activity of the Arid5a motif across conditions
Conditions sorted by the z-value of the Arid5a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
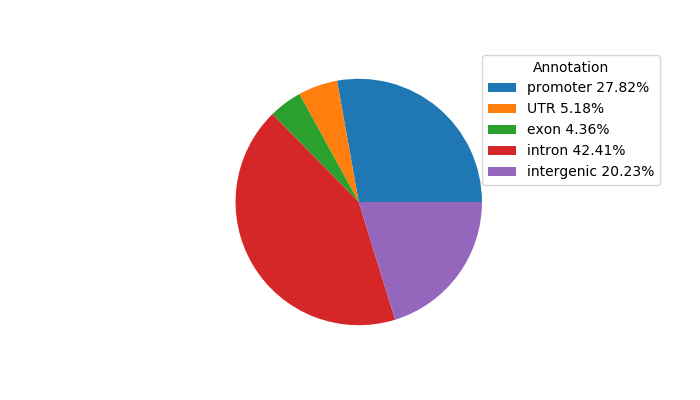
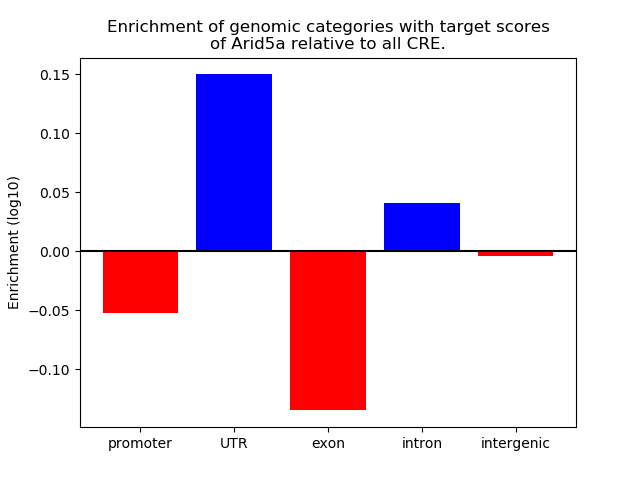
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_109928298_109928867 | 0.48 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
1738 |
0.46 |
| chr12_89814222_89814458 | 0.46 |
Nrxn3 |
neurexin III |
1857 |
0.51 |
| chr11_85896772_85897249 | 0.44 |
Tbx4 |
T-box 4 |
245 |
0.92 |
| chr17_10306194_10306345 | 0.40 |
Qk |
quaking |
13092 |
0.23 |
| chr3_141466933_141467846 | 0.38 |
Unc5c |
unc-5 netrin receptor C |
1718 |
0.37 |
| chr8_80482917_80483286 | 0.36 |
Gypa |
glycophorin A |
10680 |
0.24 |
| chr5_119838189_119838772 | 0.35 |
Tbx5 |
T-box 5 |
2325 |
0.27 |
| chr11_85887314_85887643 | 0.34 |
Tbx4 |
T-box 4 |
1056 |
0.49 |
| chr5_44917287_44917766 | 0.33 |
4930431F12Rik |
RIKEN cDNA 4930431F12 gene |
48674 |
0.12 |
| chr7_112620611_112621211 | 0.33 |
Gm24154 |
predicted gene, 24154 |
2686 |
0.25 |
| chr12_69502060_69503102 | 0.32 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
27031 |
0.14 |
| chr2_170075358_170075771 | 0.32 |
Zfp217 |
zinc finger protein 217 |
55656 |
0.15 |
| chr13_63242151_63243167 | 0.31 |
Gm47586 |
predicted gene, 47586 |
1562 |
0.22 |
| chr14_46145673_46145824 | 0.31 |
Ubb-ps |
ubiquitin B, pseudogene |
61620 |
0.11 |
| chr3_114034682_114035216 | 0.30 |
Col11a1 |
collagen, type XI, alpha 1 |
4315 |
0.36 |
| chr8_25911399_25912280 | 0.30 |
Kcnu1 |
potassium channel, subfamily U, member 1 |
147 |
0.95 |
| chr18_69834565_69834949 | 0.29 |
Gm50215 |
predicted gene, 50215 |
75967 |
0.1 |
| chr3_12382689_12382840 | 0.29 |
Gm37996 |
predicted gene, 37996 |
1845 |
0.53 |
| chr2_74670628_74670804 | 0.28 |
Hoxd13 |
homeobox D13 |
2406 |
0.09 |
| chr9_41474743_41475732 | 0.28 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
177 |
0.72 |
| chr2_103631442_103631593 | 0.27 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
65207 |
0.09 |
| chr5_43516541_43517216 | 0.25 |
C1qtnf7 |
C1q and tumor necrosis factor related protein 7 |
1116 |
0.45 |
| chr2_40295414_40295565 | 0.25 |
Gm24467 |
predicted gene, 24467 |
3028 |
0.3 |
| chr12_35533032_35533561 | 0.23 |
Gm48236 |
predicted gene, 48236 |
1125 |
0.37 |
| chr2_74707355_74707963 | 0.23 |
Hoxd8 |
homeobox D8 |
2507 |
0.08 |
| chr8_115701575_115702951 | 0.23 |
Gm15655 |
predicted gene 15655 |
2815 |
0.3 |
| chr3_132090205_132090446 | 0.22 |
Dkk2 |
dickkopf WNT signaling pathway inhibitor 2 |
5033 |
0.26 |
| chr1_177255088_177255239 | 0.22 |
Akt3 |
thymoma viral proto-oncogene 3 |
2429 |
0.26 |
| chr5_9723538_9723989 | 0.22 |
Grm3 |
glutamate receptor, metabotropic 3 |
1407 |
0.48 |
| chr3_57386472_57386752 | 0.21 |
Gm5276 |
predicted gene 5276 |
24224 |
0.18 |
| chr14_110753127_110753447 | 0.21 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
1862 |
0.32 |
| chr5_98184068_98184753 | 0.21 |
A730035I17Rik |
RIKEN cDNA A730035I17 gene |
800 |
0.59 |
| chr14_31779989_31780462 | 0.20 |
Ankrd28 |
ankyrin repeat domain 28 |
168 |
0.95 |
| chr6_147261988_147262635 | 0.20 |
Pthlh |
parathyroid hormone-like peptide |
1856 |
0.31 |
| chr14_90288772_90288923 | 0.20 |
Gm25495 |
predicted gene, 25495 |
67540 |
0.14 |
| chr2_110362115_110362868 | 0.19 |
Fibin |
fin bud initiation factor homolog (zebrafish) |
692 |
0.74 |
| chr12_41483283_41485192 | 0.19 |
Lrrn3 |
leucine rich repeat protein 3, neuronal |
2194 |
0.36 |
| chr2_33717013_33717649 | 0.19 |
9430024E24Rik |
RIKEN cDNA 9430024E24 gene |
1568 |
0.4 |
| chr13_74351957_74352108 | 0.19 |
Ccdc127 |
coiled-coil domain containing 127 |
1667 |
0.18 |
| chr2_22626612_22626763 | 0.18 |
Gad2 |
glutamic acid decarboxylase 2 |
3383 |
0.17 |
| chr6_87333037_87333530 | 0.18 |
Gm44414 |
predicted gene, 44414 |
1577 |
0.23 |
| chr18_45561882_45562417 | 0.18 |
Kcnn2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
1995 |
0.32 |
| chr18_22343502_22344104 | 0.18 |
Asxl3 |
additional sex combs like 3, transcriptional regulator |
443 |
0.91 |
| chr5_135833107_135833258 | 0.18 |
Gm23268 |
predicted gene, 23268 |
4190 |
0.13 |
| chr9_79716645_79717059 | 0.18 |
Col12a1 |
collagen, type XII, alpha 1 |
1642 |
0.36 |
| chr4_82501450_82502014 | 0.18 |
Nfib |
nuclear factor I/B |
2416 |
0.3 |
| chr2_110360491_110361293 | 0.18 |
Fibin |
fin bud initiation factor homolog (zebrafish) |
2291 |
0.34 |
| chr15_94045386_94045586 | 0.17 |
Gm30564 |
predicted gene, 30564 |
80778 |
0.1 |
| chrX_106839224_106840351 | 0.17 |
Rtl3 |
retrotransposon Gag like 3 |
857 |
0.6 |
| chr10_85183402_85183765 | 0.17 |
Cry1 |
cryptochrome 1 (photolyase-like) |
1481 |
0.42 |
| chr2_84422956_84423235 | 0.17 |
Calcrl |
calcitonin receptor-like |
2171 |
0.3 |
| chr8_41051120_41051494 | 0.17 |
Gm16193 |
predicted gene 16193 |
1803 |
0.28 |
| chr3_141981181_141982345 | 0.16 |
Bmpr1b |
bone morphogenetic protein receptor, type 1B |
501 |
0.88 |
| chr12_37112722_37113009 | 0.16 |
Meox2 |
mesenchyme homeobox 2 |
4325 |
0.22 |
| chr17_62784126_62785184 | 0.16 |
Efna5 |
ephrin A5 |
96489 |
0.09 |
| chr7_62381697_62381848 | 0.16 |
Magel2 |
melanoma antigen, family L, 2 |
4762 |
0.19 |
| chr8_115702967_115703303 | 0.16 |
Gm15655 |
predicted gene 15655 |
1943 |
0.37 |
| chr10_38968014_38968667 | 0.15 |
Lama4 |
laminin, alpha 4 |
2668 |
0.33 |
| chr5_29290255_29290406 | 0.15 |
Lmbr1 |
limb region 1 |
1884 |
0.31 |
| chr6_52230473_52231121 | 0.15 |
Hoxa9 |
homeobox A9 |
265 |
0.62 |
| chr12_71718393_71718573 | 0.15 |
Gm47555 |
predicted gene, 47555 |
35902 |
0.17 |
| chr1_185996755_185996906 | 0.15 |
Gm38268 |
predicted gene, 38268 |
115543 |
0.05 |
| chrX_114040346_114040497 | 0.15 |
Gm14896 |
predicted gene 14896 |
3894 |
0.24 |
| chr11_111605019_111605670 | 0.15 |
Gm11676 |
predicted gene 11676 |
7962 |
0.32 |
| chr19_15996837_15997609 | 0.15 |
C130060C02Rik |
RIKEN cDNA C130060C02 gene |
12145 |
0.16 |
| chr12_72233044_72233195 | 0.15 |
Rtn1 |
reticulon 1 |
2620 |
0.32 |
| chr2_6873585_6874006 | 0.15 |
Celf2 |
CUGBP, Elav-like family member 2 |
1198 |
0.49 |
| chr6_109505424_109505595 | 0.14 |
Gm44161 |
predicted gene, 44161 |
44862 |
0.17 |
| chr7_29818629_29818973 | 0.14 |
Zfp790 |
zinc finger protein 790 |
190 |
0.88 |
| chrX_134648334_134648537 | 0.14 |
Gm7855 |
predicted gene 7855 |
22691 |
0.09 |
| chr2_121804825_121804976 | 0.14 |
Frmd5 |
FERM domain containing 5 |
1972 |
0.32 |
| chr10_90578974_90579573 | 0.14 |
Anks1b |
ankyrin repeat and sterile alpha motif domain containing 1B |
2281 |
0.42 |
| chr17_53463562_53463837 | 0.14 |
Efhb |
EF hand domain family, member B |
378 |
0.81 |
| chr9_94535439_94536002 | 0.14 |
Dipk2a |
divergent protein kinase domain 2A |
2361 |
0.35 |
| chr5_25529088_25529591 | 0.14 |
1700096K18Rik |
RIKEN cDNA 1700096K18 gene |
674 |
0.58 |
| chr7_84899042_84899193 | 0.14 |
Olfr290 |
olfactory receptor 290 |
13958 |
0.12 |
| chr4_29133351_29134210 | 0.14 |
Gm11918 |
predicted gene 11918 |
7834 |
0.23 |
| chr2_74741953_74742174 | 0.14 |
Hoxd3 |
homeobox D3 |
2444 |
0.1 |
| chr6_135066090_135066467 | 0.14 |
Gprc5a |
G protein-coupled receptor, family C, group 5, member A |
627 |
0.67 |
| chr4_119816287_119816915 | 0.14 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
2106 |
0.42 |
| chr1_178317819_178318084 | 0.14 |
B230369F24Rik |
RIKEN cDNA B230369F24 gene |
598 |
0.52 |
| chr10_59325085_59325236 | 0.13 |
P4ha1 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
1792 |
0.38 |
| chr10_109007772_109008002 | 0.13 |
Syt1 |
synaptotagmin I |
1213 |
0.6 |
| chr9_71894609_71895239 | 0.13 |
Tcf12 |
transcription factor 12 |
1061 |
0.37 |
| chr5_30281813_30282015 | 0.13 |
Drc1 |
dynein regulatory complex subunit 1 |
526 |
0.74 |
| chr1_138244489_138245049 | 0.13 |
1700113B19Rik |
RIKEN cDNA 1700113B19 gene |
16873 |
0.15 |
| chr2_157025463_157025614 | 0.13 |
Soga1 |
suppressor of glucose, autophagy associated 1 |
1934 |
0.23 |
| chr12_46813397_46813578 | 0.13 |
Gm48542 |
predicted gene, 48542 |
1979 |
0.34 |
| chr8_114884771_114884922 | 0.13 |
Gm22556 |
predicted gene, 22556 |
168067 |
0.03 |
| chr13_109443973_109444887 | 0.13 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
2247 |
0.46 |
| chr4_84746085_84746236 | 0.13 |
Gm12415 |
predicted gene 12415 |
4685 |
0.27 |
| chr2_150115282_150115433 | 0.13 |
Gm14140 |
predicted gene 14140 |
3781 |
0.22 |
| chr8_120232089_120233564 | 0.13 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
4370 |
0.19 |
| chr6_52233543_52234485 | 0.13 |
Hoxa10 |
homeobox A10 |
690 |
0.34 |
| chr9_43069907_43070263 | 0.12 |
Arhgef12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
27453 |
0.17 |
| chr10_107882911_107883105 | 0.12 |
Otogl |
otogelin-like |
29126 |
0.22 |
| chr12_99756799_99758054 | 0.12 |
Gm47208 |
predicted gene, 47208 |
4828 |
0.2 |
| chr8_90956080_90956960 | 0.12 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
1085 |
0.49 |
| chr5_17848306_17848609 | 0.12 |
Cd36 |
CD36 molecule |
1272 |
0.61 |
| chr13_65276016_65276167 | 0.12 |
Zfp369 |
zinc finger protein 369 |
2723 |
0.11 |
| chr16_34999592_34999743 | 0.12 |
Mylk |
myosin, light polypeptide kinase |
4860 |
0.18 |
| chr6_21219551_21220038 | 0.12 |
Kcnd2 |
potassium voltage-gated channel, Shal-related family, member 2 |
4291 |
0.29 |
| chrX_109097818_109098006 | 0.12 |
Sh3bgrl |
SH3-binding domain glutamic acid-rich protein like |
2403 |
0.29 |
| chr9_75610075_75610368 | 0.12 |
Tmod2 |
tropomodulin 2 |
854 |
0.52 |
| chr3_58424450_58424601 | 0.12 |
Tsc22d2 |
TSC22 domain family, member 2 |
7041 |
0.19 |
| chr17_44813433_44813945 | 0.12 |
Runx2 |
runt related transcription factor 2 |
537 |
0.8 |
| chr16_78238213_78238675 | 0.12 |
Gm25916 |
predicted gene, 25916 |
1814 |
0.31 |
| chr2_114654081_114654668 | 0.12 |
Gm13972 |
predicted gene 13972 |
51 |
0.9 |
| chr5_117087628_117088305 | 0.12 |
Suds3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
10845 |
0.15 |
| chr1_186702387_186703121 | 0.12 |
Tgfb2 |
transforming growth factor, beta 2 |
1579 |
0.29 |
| chr12_86681553_86682494 | 0.12 |
Vash1 |
vasohibin 1 |
3323 |
0.18 |
| chr1_194623571_194625393 | 0.12 |
Plxna2 |
plexin A2 |
4657 |
0.21 |
| chr10_21998478_21999596 | 0.11 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
2446 |
0.24 |
| chr19_12518482_12518707 | 0.11 |
Gm47243 |
predicted gene, 47243 |
1206 |
0.3 |
| chr15_40725754_40725905 | 0.11 |
Zfpm2 |
zinc finger protein, multitype 2 |
60839 |
0.15 |
| chr17_84596177_84596438 | 0.11 |
Plekhh2 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
20757 |
0.12 |
| chr14_79429804_79429955 | 0.11 |
Kbtbd7 |
kelch repeat and BTB (POZ) domain containing 7 |
3368 |
0.18 |
| chr16_36983491_36983642 | 0.11 |
Fbxo40 |
F-box protein 40 |
4430 |
0.15 |
| chr13_45192224_45192456 | 0.11 |
Gm8513 |
predicted gene 8513 |
4804 |
0.23 |
| chr19_8741145_8741296 | 0.11 |
Stx5a |
syntaxin 5A |
193 |
0.62 |
| chr2_75154680_75154968 | 0.11 |
2600014E21Rik |
RIKEN cDNA 2600014E21 gene |
15944 |
0.14 |
| chr3_146681740_146682065 | 0.11 |
Samd13 |
sterile alpha motif domain containing 13 |
508 |
0.72 |
| chr3_79818328_79818717 | 0.11 |
Gm26420 |
predicted gene, 26420 |
16411 |
0.17 |
| chr11_53567066_53567387 | 0.11 |
A430108G06Rik |
RIKEN cDNA A430108G06 gene |
79 |
0.6 |
| chr13_44841435_44841934 | 0.11 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
901 |
0.61 |
| chr16_74780405_74780742 | 0.11 |
Gm49674 |
predicted gene, 49674 |
95365 |
0.08 |
| chr9_112229635_112229830 | 0.11 |
2310075C17Rik |
RIKEN cDNA 2310075C17 gene |
2153 |
0.26 |
| chr14_31778839_31778990 | 0.11 |
Ankrd28 |
ankyrin repeat domain 28 |
1479 |
0.38 |
| chr12_95699798_95699986 | 0.11 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
4535 |
0.2 |
| chr7_42373282_42373433 | 0.11 |
Vmn2r-ps59 |
vomeronasal 2, receptor, pseudogene 59 |
5102 |
0.17 |
| chr18_84082718_84083733 | 0.11 |
Tshz1 |
teashirt zinc finger family member 1 |
1850 |
0.29 |
| chr17_22716866_22717055 | 0.11 |
Gm20008 |
predicted gene, 20008 |
23 |
0.97 |
| chr7_115865614_115865765 | 0.11 |
Sox6 |
SRY (sex determining region Y)-box 6 |
5837 |
0.3 |
| chrX_95195777_95197048 | 0.11 |
Arhgef9 |
CDC42 guanine nucleotide exchange factor (GEF) 9 |
40 |
0.98 |
| chr6_92478006_92478157 | 0.10 |
Prickle2 |
prickle planar cell polarity protein 2 |
3311 |
0.34 |
| chr4_58668245_58668566 | 0.10 |
Gm12580 |
predicted gene 12580 |
9813 |
0.22 |
| chr5_14028529_14028806 | 0.10 |
Gm43519 |
predicted gene 43519 |
773 |
0.63 |
| chr5_49284738_49286021 | 0.10 |
Kcnip4 |
Kv channel interacting protein 4 |
280 |
0.92 |
| chr1_103933897_103934048 | 0.10 |
Gm37789 |
predicted gene, 37789 |
209627 |
0.03 |
| chr3_116922901_116923052 | 0.10 |
Gm42892 |
predicted gene 42892 |
21354 |
0.12 |
| chr19_46928778_46928929 | 0.10 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
372 |
0.86 |
| chr10_82152183_82152457 | 0.10 |
Gm47572 |
predicted gene, 47572 |
4781 |
0.14 |
| chr4_144896034_144896645 | 0.10 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
3120 |
0.28 |
| chr2_79087179_79088090 | 0.10 |
Gm14469 |
predicted gene 14469 |
48900 |
0.14 |
| chr1_136915480_136915909 | 0.10 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
24889 |
0.19 |
| chr1_156839730_156839989 | 0.10 |
Angptl1 |
angiopoietin-like 1 |
917 |
0.42 |
| chr8_8850795_8850946 | 0.10 |
Gm44515 |
predicted gene 44515 |
81829 |
0.07 |
| chr14_76417732_76418489 | 0.10 |
Tsc22d1 |
TSC22 domain family, member 1 |
244 |
0.95 |
| chr6_63258813_63259293 | 0.10 |
9330118I20Rik |
RIKEN cDNA 9330118I20 gene |
1428 |
0.39 |
| chr8_114592004_114592229 | 0.10 |
Gm16116 |
predicted gene 16116 |
2784 |
0.29 |
| chr19_59940546_59941181 | 0.10 |
Rab11fip2 |
RAB11 family interacting protein 2 (class I) |
2137 |
0.29 |
| chr14_110750421_110750572 | 0.10 |
Slitrk6 |
SLIT and NTRK-like family, member 6 |
4653 |
0.2 |
| chr6_21217917_21219303 | 0.10 |
Kcnd2 |
potassium voltage-gated channel, Shal-related family, member 2 |
3107 |
0.33 |
| chrX_106921813_106922604 | 0.10 |
Lpar4 |
lysophosphatidic acid receptor 4 |
1583 |
0.46 |
| chr1_147720137_147720384 | 0.10 |
Gm38284 |
predicted gene, 38284 |
204534 |
0.03 |
| chrX_110809229_110809704 | 0.09 |
Gm44593 |
predicted gene 44593 |
2858 |
0.31 |
| chr8_109904347_109904498 | 0.09 |
Gm16349 |
predicted gene 16349 |
20966 |
0.11 |
| chr14_104640336_104640571 | 0.09 |
D130009I18Rik |
RIKEN cDNA D130009I18 gene |
1309 |
0.47 |
| chr18_90002590_90002741 | 0.09 |
Gm6173 |
predicted gene 6173 |
16414 |
0.25 |
| chr4_38855115_38855266 | 0.09 |
Gm12381 |
predicted gene 12381 |
293 |
0.95 |
| chr1_94469970_94470266 | 0.09 |
Gm7895 |
predicted gene 7895 |
231 |
0.96 |
| chr6_93715774_93716464 | 0.09 |
Gm7812 |
predicted gene 7812 |
65894 |
0.11 |
| chr2_66041980_66042300 | 0.09 |
Galnt3 |
polypeptide N-acetylgalactosaminyltransferase 3 |
51931 |
0.12 |
| chr3_97903523_97903692 | 0.09 |
Sec22b |
SEC22 homolog B, vesicle trafficking protein |
2380 |
0.22 |
| chr4_49381036_49381187 | 0.09 |
Acnat2 |
acyl-coenzyme A amino acid N-acyltransferase 2 |
2465 |
0.21 |
| chr4_81566839_81567150 | 0.09 |
Gm11765 |
predicted gene 11765 |
105262 |
0.07 |
| chr2_74660173_74660324 | 0.09 |
Evx2 |
even-skipped homeobox 2 |
691 |
0.39 |
| chr5_122950311_122951097 | 0.09 |
Kdm2b |
lysine (K)-specific demethylase 2B |
1286 |
0.34 |
| chr10_89255473_89255822 | 0.09 |
Ano4 |
anoctamin 4 |
1489 |
0.56 |
| chr10_110233512_110233722 | 0.09 |
Gm47340 |
predicted gene, 47340 |
93448 |
0.08 |
| chr19_34691351_34691502 | 0.09 |
Slc16a12 |
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
121 |
0.95 |
| chr12_50122094_50122289 | 0.09 |
Gm40418 |
predicted gene, 40418 |
1882 |
0.51 |
| chr10_121296462_121296721 | 0.09 |
Tbc1d30 |
TBC1 domain family, member 30 |
444 |
0.8 |
| chr10_21993256_21993468 | 0.09 |
Sgk1 |
serum/glucocorticoid regulated kinase 1 |
528 |
0.75 |
| chr2_82055274_82055801 | 0.09 |
Zfp804a |
zinc finger protein 804A |
2315 |
0.43 |
| chr10_75214046_75214484 | 0.09 |
Specc1l |
sperm antigen with calponin homology and coiled-coil domains 1-like |
1845 |
0.36 |
| chr17_36181694_36181845 | 0.09 |
Gm20495 |
predicted gene 20495 |
661 |
0.31 |
| chr10_96744086_96744687 | 0.09 |
Gm6859 |
predicted gene 6859 |
25442 |
0.15 |
| chr2_172731809_172732445 | 0.09 |
Gm22773 |
predicted gene, 22773 |
132214 |
0.04 |
| chr15_15755741_15755892 | 0.09 |
4930557F08Rik |
RIKEN cDNA 4930557F08 gene |
24178 |
0.24 |
| chr5_131836026_131836207 | 0.09 |
4930563F08Rik |
RIKEN cDNA 4930563F08 gene |
44153 |
0.11 |
| chr7_110859157_110859397 | 0.09 |
Lyve1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
3676 |
0.19 |
| chr2_92601517_92601703 | 0.09 |
Chst1 |
carbohydrate sulfotransferase 1 |
1903 |
0.26 |
| chr10_33621658_33622333 | 0.09 |
Gm47911 |
predicted gene, 47911 |
286 |
0.85 |
| chr5_65133569_65133946 | 0.09 |
Klhl5 |
kelch-like 5 |
2086 |
0.25 |
| chr7_137302193_137302490 | 0.09 |
Ebf3 |
early B cell factor 3 |
11575 |
0.19 |
| chrX_12936850_12937769 | 0.09 |
AA414768 |
expressed sequence AA414768 |
437 |
0.81 |
| chrX_89558260_89558411 | 0.09 |
Gm6973 |
predicted gene 6973 |
117440 |
0.05 |
| chr8_12915306_12916734 | 0.09 |
Mcf2l |
mcf.2 transforming sequence-like |
45 |
0.76 |
| chr8_73340046_73340197 | 0.09 |
Large1 |
LARGE xylosyl- and glucuronyltransferase 1 |
12435 |
0.28 |
| chr17_47141488_47142433 | 0.08 |
Trerf1 |
transcriptional regulating factor 1 |
744 |
0.72 |
| chr18_81471682_81471945 | 0.08 |
Gm50412 |
predicted gene, 50412 |
8544 |
0.23 |
| chr13_63245084_63245617 | 0.08 |
Gm47586 |
predicted gene, 47586 |
1129 |
0.31 |
| chr17_64329718_64329909 | 0.08 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
2052 |
0.38 |
| chr18_70804952_70805557 | 0.08 |
Gm21046 |
predicted gene, 21046 |
23367 |
0.2 |
| chr18_3510795_3510946 | 0.08 |
Bambi |
BMP and activin membrane-bound inhibitor |
2947 |
0.21 |
| chr9_52676918_52677560 | 0.08 |
AI593442 |
expressed sequence AI593442 |
2190 |
0.3 |
| chr2_37519004_37519477 | 0.08 |
Gpr21 |
G protein-coupled receptor 21 |
2614 |
0.2 |
| chr13_104228230_104228763 | 0.08 |
Cenpk |
centromere protein K |
115 |
0.71 |
| chr8_71602124_71603477 | 0.08 |
Fam129c |
family with sequence similarity 129, member C |
449 |
0.63 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.1 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0044688 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |