Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
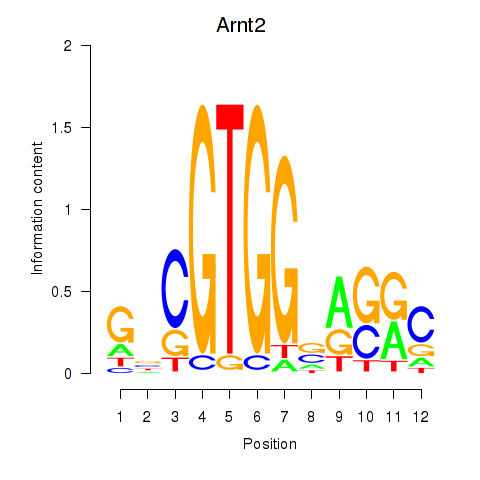
Results for Arnt2
Z-value: 0.16
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.8 | aryl hydrocarbon receptor nuclear translocator 2 |
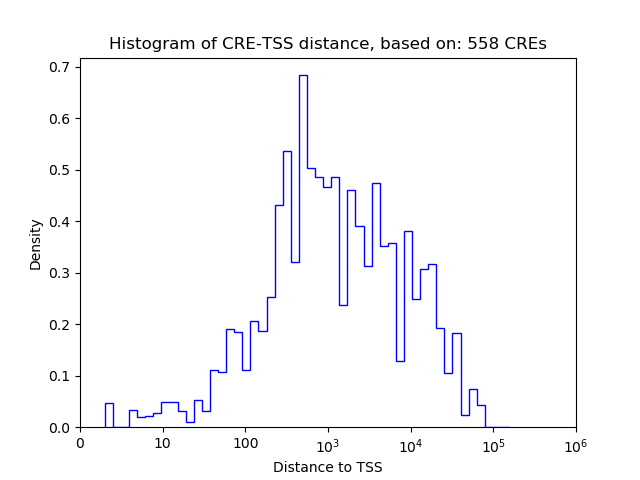
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_84372196_84372536 | Arnt2 | 129 | 0.959493 | -0.44 | 4.1e-04 | Click! |
| chr7_84409381_84409909 | Arnt2 | 238 | 0.919535 | -0.41 | 1.1e-03 | Click! |
| chr7_84371835_84371997 | Arnt2 | 579 | 0.734859 | -0.33 | 9.8e-03 | Click! |
| chr7_84409925_84410547 | Arnt2 | 60 | 0.971569 | -0.30 | 2.1e-02 | Click! |
| chr7_84407791_84408163 | Arnt2 | 1906 | 0.307516 | -0.29 | 2.2e-02 | Click! |
Activity of the Arnt2 motif across conditions
Conditions sorted by the z-value of the Arnt2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
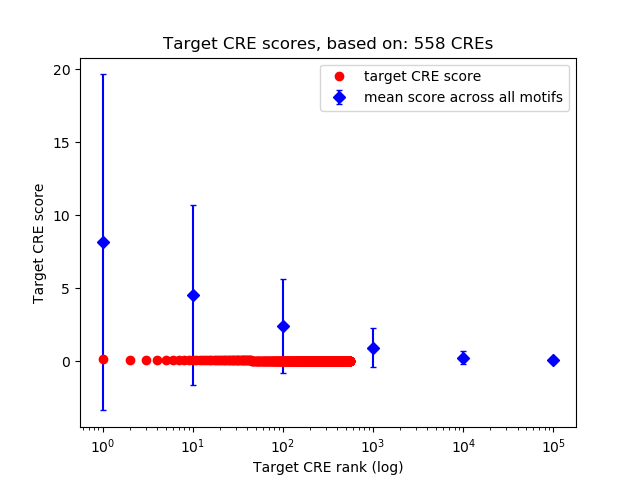
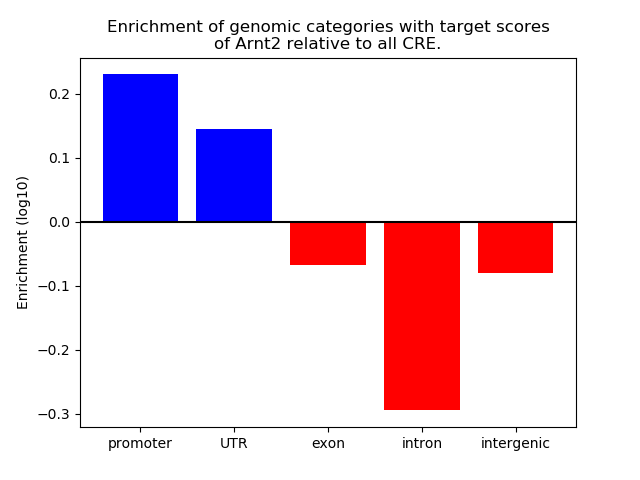
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_105125289_105128976 | 0.18 |
Wt1 |
Wilms tumor 1 homolog |
78 |
0.91 |
| chr14_63370712_63371238 | 0.11 |
Blk |
B lymphoid kinase |
5443 |
0.18 |
| chr18_56470317_56470937 | 0.11 |
Gramd3 |
GRAM domain containing 3 |
1360 |
0.42 |
| chr7_16885022_16886331 | 0.11 |
2700080J24Rik |
RIKEN cDNA 2700080J24 gene |
5391 |
0.09 |
| chr3_96458110_96459469 | 0.11 |
Gm10685 |
predicted gene 10685 |
580 |
0.4 |
| chrX_137118132_137120673 | 0.10 |
Esx1 |
extraembryonic, spermatogenesis, homeobox 1 |
769 |
0.36 |
| chr5_100570574_100571670 | 0.10 |
Plac8 |
placenta-specific 8 |
1070 |
0.43 |
| chr13_73262153_73264451 | 0.10 |
Irx4 |
Iroquois homeobox 4 |
2805 |
0.22 |
| chr3_129216664_129219042 | 0.09 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
3578 |
0.2 |
| chr13_48664673_48666478 | 0.09 |
Barx1 |
BarH-like homeobox 1 |
2577 |
0.26 |
| chr4_134864478_134865098 | 0.09 |
Rhd |
Rh blood group, D antigen |
252 |
0.91 |
| chr2_170130220_170131144 | 0.09 |
Zfp217 |
zinc finger protein 217 |
538 |
0.86 |
| chr7_44918454_44918709 | 0.08 |
Ap2a1 |
adaptor-related protein complex 2, alpha 1 subunit |
1847 |
0.14 |
| chr2_167688724_167690153 | 0.08 |
Tmem189 |
transmembrane protein 189 |
384 |
0.52 |
| chr3_96450951_96451691 | 0.08 |
Gm26232 |
predicted gene, 26232 |
875 |
0.23 |
| chr2_91949326_91950727 | 0.07 |
Dgkz |
diacylglycerol kinase zeta |
285 |
0.84 |
| chr17_57233601_57234302 | 0.07 |
C3 |
complement component 3 |
5815 |
0.11 |
| chr11_116572166_116572368 | 0.07 |
Ube2o |
ubiquitin-conjugating enzyme E2O |
9180 |
0.1 |
| chr5_117135404_117135836 | 0.07 |
Taok3 |
TAO kinase 3 |
1979 |
0.25 |
| chr16_38362450_38363226 | 0.07 |
Popdc2 |
popeye domain containing 2 |
593 |
0.63 |
| chr7_120173917_120175138 | 0.07 |
Anks4b |
ankyrin repeat and sterile alpha motif domain containing 4B |
669 |
0.61 |
| chr14_54626108_54626259 | 0.07 |
Psmb11 |
proteasome (prosome, macropain) subunit, beta type, 11 |
873 |
0.32 |
| chr14_115040989_115042168 | 0.07 |
Mir17hg |
Mir17 host gene (non-protein coding) |
1301 |
0.2 |
| chr1_71938036_71938516 | 0.07 |
Gm28818 |
predicted gene 28818 |
21151 |
0.17 |
| chr2_148043571_148045987 | 0.07 |
Foxa2 |
forkhead box A2 |
685 |
0.65 |
| chr18_60723029_60723795 | 0.07 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
4151 |
0.19 |
| chr5_107724563_107727169 | 0.07 |
Gfi1 |
growth factor independent 1 transcription repressor |
61 |
0.88 |
| chr5_66080287_66081072 | 0.07 |
Rbm47 |
RNA binding motif protein 47 |
305 |
0.84 |
| chr1_184731287_184732512 | 0.06 |
Hlx |
H2.0-like homeobox |
301 |
0.86 |
| chr5_119669544_119672401 | 0.06 |
Tbx3 |
T-box 3 |
46 |
0.85 |
| chr4_148924034_148924185 | 0.06 |
Casz1 |
castor zinc finger 1 |
9005 |
0.16 |
| chr11_75463013_75463425 | 0.06 |
Mir22 |
microRNA 22 |
497 |
0.52 |
| chr2_118702607_118704323 | 0.06 |
Ankrd63 |
ankyrin repeat domain 63 |
498 |
0.71 |
| chr10_68156328_68157069 | 0.06 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
20072 |
0.24 |
| chrX_167227841_167227992 | 0.06 |
Tmsb4x |
thymosin, beta 4, X chromosome |
18601 |
0.18 |
| chr10_63062957_63063108 | 0.06 |
Pbld1 |
phenazine biosynthesis-like protein domain containing 1 |
772 |
0.45 |
| chr16_17914132_17915323 | 0.06 |
Gsc2 |
goosecoid homebox 2 |
332 |
0.74 |
| chr9_72409899_72410132 | 0.06 |
BC065403 |
cDNA sequence BC065403 |
70 |
0.89 |
| chr16_94086633_94087964 | 0.06 |
Sim2 |
single-minded family bHLH transcription factor 2 |
2367 |
0.22 |
| chr2_25094719_25095656 | 0.06 |
Noxa1 |
NADPH oxidase activator 1 |
38 |
0.94 |
| chr11_102145120_102148094 | 0.06 |
Nags |
N-acetylglutamate synthase |
241 |
0.58 |
| chr11_96938596_96938782 | 0.06 |
Pnpo |
pyridoxine 5'-phosphate oxidase |
5090 |
0.09 |
| chr15_96465106_96465417 | 0.05 |
Scaf11 |
SR-related CTD-associated factor 11 |
4418 |
0.23 |
| chr1_191894497_191894858 | 0.05 |
1700034H15Rik |
RIKEN cDNA 1700034H15 gene |
4452 |
0.17 |
| chr4_110227524_110228376 | 0.05 |
Elavl4 |
ELAV like RNA binding protein 4 |
4802 |
0.34 |
| chr5_103769379_103770425 | 0.05 |
Aff1 |
AF4/FMR2 family, member 1 |
14457 |
0.2 |
| chr10_50891809_50893729 | 0.05 |
Sim1 |
single-minded family bHLH transcription factor 1 |
1985 |
0.43 |
| chr12_84408189_84409305 | 0.05 |
Entpd5 |
ectonucleoside triphosphate diphosphohydrolase 5 |
237 |
0.57 |
| chr3_21987826_21988387 | 0.05 |
Gm43674 |
predicted gene 43674 |
10362 |
0.21 |
| chr7_31115037_31115979 | 0.05 |
Hpn |
hepsin |
218 |
0.86 |
| chr6_138171729_138171880 | 0.05 |
Mgst1 |
microsomal glutathione S-transferase 1 |
28950 |
0.23 |
| chr19_6854909_6856205 | 0.05 |
Ccdc88b |
coiled-coil domain containing 88B |
319 |
0.8 |
| chr7_137321110_137323113 | 0.05 |
Ebf3 |
early B cell factor 3 |
7666 |
0.19 |
| chr11_97415560_97416471 | 0.05 |
Arhgap23 |
Rho GTPase activating protein 23 |
482 |
0.77 |
| chr4_44711553_44714511 | 0.05 |
Pax5 |
paired box 5 |
2545 |
0.22 |
| chr15_78571734_78572724 | 0.05 |
Rac2 |
Rac family small GTPase 2 |
552 |
0.62 |
| chr15_8279541_8279705 | 0.05 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
32520 |
0.18 |
| chr18_80468808_80469915 | 0.05 |
Ctdp1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
334 |
0.62 |
| chr6_4903006_4904131 | 0.05 |
Ppp1r9a |
protein phosphatase 1, regulatory subunit 9A |
136 |
0.95 |
| chr12_86881882_86884741 | 0.05 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
1487 |
0.4 |
| chr12_84218014_84219177 | 0.04 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
286 |
0.83 |
| chr12_56533030_56535358 | 0.04 |
Nkx2-1 |
NK2 homeobox 1 |
912 |
0.36 |
| chr14_121137043_121138056 | 0.04 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
35470 |
0.2 |
| chr5_104044164_104044315 | 0.04 |
Nudt9 |
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
2067 |
0.18 |
| chr3_51727954_51728653 | 0.04 |
Gm37342 |
predicted gene, 37342 |
33785 |
0.1 |
| chr19_10015065_10016667 | 0.04 |
Rab3il1 |
RAB3A interacting protein (rabin3)-like 1 |
822 |
0.48 |
| chr5_124452958_124453201 | 0.04 |
Kmt5a |
lysine methyltransferase 5A |
1908 |
0.18 |
| chr19_46304366_46306224 | 0.04 |
4833438C02Rik |
RIKEN cDNA 4833438C02 gene |
310 |
0.52 |
| chr14_118987522_118988155 | 0.04 |
Gm49768 |
predicted gene, 49768 |
4078 |
0.21 |
| chr2_154551905_154553077 | 0.04 |
Actl10 |
actin-like 10 |
715 |
0.51 |
| chr12_102368568_102368815 | 0.04 |
Rin3 |
Ras and Rab interactor 3 |
13911 |
0.19 |
| chr2_30995683_30996883 | 0.04 |
Usp20 |
ubiquitin specific peptidase 20 |
235 |
0.9 |
| chrX_75842884_75843270 | 0.04 |
Pls3 |
plastin 3 (T-isoform) |
14 |
0.98 |
| chr3_129213621_129215189 | 0.04 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
130 |
0.95 |
| chr6_97311036_97312726 | 0.04 |
Frmd4b |
FERM domain containing 4B |
499 |
0.83 |
| chr1_61376358_61376610 | 0.04 |
9530026F06Rik |
RIKEN cDNA 9530026F06 gene |
1948 |
0.35 |
| chr5_75152837_75154692 | 0.04 |
Gm42802 |
predicted gene 42802 |
111 |
0.58 |
| chr4_45972347_45973304 | 0.04 |
Tdrd7 |
tudor domain containing 7 |
9 |
0.98 |
| chr1_191184645_191186177 | 0.04 |
Atf3 |
activating transcription factor 3 |
2071 |
0.24 |
| chr2_101649005_101649668 | 0.04 |
Rag1 |
recombination activating 1 |
165 |
0.54 |
| chr7_45923986_45924770 | 0.04 |
Ccdc114 |
coiled-coil domain containing 114 |
135 |
0.57 |
| chr15_93513841_93514524 | 0.04 |
Prickle1 |
prickle planar cell polarity protein 1 |
5341 |
0.28 |
| chr9_50751156_50752468 | 0.04 |
Cryab |
crystallin, alpha B |
65 |
0.86 |
| chrX_164257385_164257820 | 0.04 |
Bmx |
BMX non-receptor tyrosine kinase |
591 |
0.69 |
| chr10_127177539_127178955 | 0.04 |
Slc26a10 |
solute carrier family 26, member 10 |
1258 |
0.23 |
| chr12_86974337_86975184 | 0.04 |
Zdhhc22 |
zinc finger, DHHC-type containing 22 |
9075 |
0.15 |
| chr14_70993754_70994386 | 0.04 |
Gfra2 |
glial cell line derived neurotrophic factor family receptor alpha 2 |
52184 |
0.15 |
| chr7_44553651_44553839 | 0.04 |
Nr1h2 |
nuclear receptor subfamily 1, group H, member 2 |
37 |
0.92 |
| chr7_100911201_100912201 | 0.04 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
17011 |
0.13 |
| chr8_11476446_11477451 | 0.04 |
E230013L22Rik |
RIKEN cDNA E230013L22 gene |
981 |
0.37 |
| chr10_119011332_119011760 | 0.04 |
Gm47461 |
predicted gene, 47461 |
10124 |
0.17 |
| chr2_91083412_91083784 | 0.04 |
Spi1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
1190 |
0.32 |
| chr5_32139282_32139946 | 0.04 |
Fosl2 |
fos-like antigen 2 |
3441 |
0.19 |
| chr8_122546551_122549259 | 0.04 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
3424 |
0.12 |
| chr7_25787527_25787868 | 0.04 |
Axl |
AXL receptor tyrosine kinase |
125 |
0.92 |
| chr9_59660933_59662180 | 0.04 |
Pkm |
pyruvate kinase, muscle |
3246 |
0.17 |
| chr10_127515904_127517428 | 0.04 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
1699 |
0.19 |
| chr2_37793046_37793898 | 0.03 |
Crb2 |
crumbs family member 2 |
1697 |
0.41 |
| chr9_32408669_32409941 | 0.03 |
Kcnj1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
15320 |
0.15 |
| chr14_60734968_60735562 | 0.03 |
Spata13 |
spermatogenesis associated 13 |
2359 |
0.3 |
| chr5_147758103_147758896 | 0.03 |
Gm43156 |
predicted gene 43156 |
12983 |
0.19 |
| chr11_58999701_58999943 | 0.03 |
Obscn |
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
537 |
0.56 |
| chr11_100930890_100931041 | 0.03 |
Stat3 |
signal transducer and activator of transcription 3 |
8415 |
0.14 |
| chr8_84711904_84712777 | 0.03 |
Nfix |
nuclear factor I/X |
1728 |
0.2 |
| chr2_156779787_156779956 | 0.03 |
Myl9 |
myosin, light polypeptide 9, regulatory |
1354 |
0.3 |
| chr3_68676477_68677404 | 0.03 |
Il12a |
interleukin 12a |
13704 |
0.19 |
| chr11_85831867_85832577 | 0.03 |
2610027K06Rik |
RIKEN cDNA 2610027K06 gene |
1 |
0.83 |
| chr8_126474184_126475045 | 0.03 |
Tarbp1 |
TAR RNA binding protein 1 |
451 |
0.79 |
| chr1_132381900_132382379 | 0.03 |
Gm15849 |
predicted gene 15849 |
1010 |
0.43 |
| chr7_44524691_44525693 | 0.03 |
Mybpc2 |
myosin binding protein C, fast-type |
536 |
0.49 |
| chr9_44509311_44510671 | 0.03 |
Bcl9l |
B cell CLL/lymphoma 9-like |
10719 |
0.07 |
| chr12_51709017_51709206 | 0.03 |
Strn3 |
striatin, calmodulin binding protein 3 |
17214 |
0.16 |
| chr7_49527363_49527925 | 0.03 |
Nav2 |
neuron navigator 2 |
20548 |
0.23 |
| chr2_29750670_29752031 | 0.03 |
Gm13420 |
predicted gene 13420 |
335 |
0.8 |
| chr5_30906078_30906273 | 0.03 |
Ost4 |
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
1550 |
0.19 |
| chr2_152636550_152636849 | 0.03 |
Rem1 |
rad and gem related GTP binding protein 1 |
3732 |
0.1 |
| chr12_111815824_111816910 | 0.03 |
Zfyve21 |
zinc finger, FYVE domain containing 21 |
111 |
0.94 |
| chr5_111519326_111519818 | 0.03 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
61850 |
0.1 |
| chr14_27114231_27115583 | 0.03 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
8 |
0.98 |
| chr14_61288659_61289212 | 0.03 |
Gm19097 |
predicted gene, 19097 |
4147 |
0.15 |
| chr14_76150578_76150729 | 0.03 |
Nufip1 |
nuclear fragile X mental retardation protein interacting protein 1 |
39762 |
0.14 |
| chr7_111081451_111082886 | 0.03 |
Eif4g2 |
eukaryotic translation initiation factor 4, gamma 2 |
27 |
0.97 |
| chr2_32380493_32380743 | 0.03 |
1110008P14Rik |
RIKEN cDNA 1110008P14 gene |
1244 |
0.25 |
| chr6_99845386_99845693 | 0.03 |
Gm6681 |
predicted gene 6681 |
5225 |
0.2 |
| chr14_70622289_70622914 | 0.03 |
Dmtn |
dematin actin binding protein |
3554 |
0.14 |
| chr6_88116254_88117368 | 0.03 |
Mir6376 |
microRNA 6376 |
12553 |
0.13 |
| chr17_56803471_56803622 | 0.03 |
Rfx2 |
regulatory factor X, 2 (influences HLA class II expression) |
19842 |
0.1 |
| chr3_87761429_87761580 | 0.03 |
Pear1 |
platelet endothelial aggregation receptor 1 |
710 |
0.6 |
| chr11_98392512_98393112 | 0.03 |
Pgap3 |
post-GPI attachment to proteins 3 |
740 |
0.42 |
| chr3_96349456_96350053 | 0.03 |
Platr30 |
pluripotency associated transcript 30 |
173 |
0.84 |
| chr10_43478627_43479241 | 0.03 |
Bend3 |
BEN domain containing 3 |
103 |
0.95 |
| chr7_118490935_118492121 | 0.03 |
Itpripl2 |
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
447 |
0.78 |
| chr10_79978352_79978697 | 0.03 |
Tmem259 |
transmembrane protein 259 |
249 |
0.78 |
| chr9_35331893_35332155 | 0.03 |
Gm33838 |
predicted gene, 33838 |
24425 |
0.11 |
| chr10_118867942_118869119 | 0.03 |
Dyrk2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
231 |
0.89 |
| chr2_145787355_145788162 | 0.03 |
Rin2 |
Ras and Rab interactor 2 |
1596 |
0.46 |
| chr5_137644960_137646061 | 0.03 |
Irs3 |
insulin receptor substrate 3 |
27 |
0.93 |
| chr8_124750940_124751848 | 0.03 |
Fam89a |
family with sequence similarity 89, member A |
438 |
0.74 |
| chr19_23758400_23759783 | 0.03 |
Apba1 |
amyloid beta (A4) precursor protein binding, family A, member 1 |
204 |
0.93 |
| chr12_105008802_105010413 | 0.03 |
Syne3 |
spectrin repeat containing, nuclear envelope family member 3 |
202 |
0.89 |
| chr17_56510067_56512183 | 0.03 |
Znrf4 |
zinc and ring finger 4 |
1306 |
0.36 |
| chr12_109455257_109457986 | 0.03 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
2426 |
0.16 |
| chr15_35302239_35303258 | 0.03 |
Osr2 |
odd-skipped related 2 |
2448 |
0.29 |
| chr7_25243630_25243781 | 0.03 |
Gsk3a |
glycogen synthase kinase 3 alpha |
5854 |
0.08 |
| chr11_114468392_114468903 | 0.03 |
4932435O22Rik |
RIKEN cDNA 4932435O22 gene |
19787 |
0.23 |
| chr11_103364463_103364614 | 0.03 |
Arhgap27 |
Rho GTPase activating protein 27 |
846 |
0.41 |
| chr15_81585550_81586926 | 0.03 |
Ep300 |
E1A binding protein p300 |
18 |
0.58 |
| chr10_20204894_20205970 | 0.03 |
Rpl32l |
ribosomal protein L32-like |
3058 |
0.2 |
| chr4_88031567_88032752 | 0.03 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
654 |
0.59 |
| chr14_62342534_62343111 | 0.03 |
Rnaseh2b |
ribonuclease H2, subunit B |
4477 |
0.25 |
| chr12_80111226_80112041 | 0.03 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
1361 |
0.31 |
| chr2_119742307_119743100 | 0.03 |
Itpka |
inositol 1,4,5-trisphosphate 3-kinase A |
366 |
0.74 |
| chr10_63276115_63276288 | 0.03 |
Herc4 |
hect domain and RLD 4 |
2202 |
0.2 |
| chr1_55026194_55027366 | 0.03 |
Sf3b1 |
splicing factor 3b, subunit 1 |
593 |
0.66 |
| chr10_127169955_127171248 | 0.03 |
Slc26a10 |
solute carrier family 26, member 10 |
4512 |
0.09 |
| chr4_123961418_123961932 | 0.03 |
Gm12902 |
predicted gene 12902 |
35441 |
0.11 |
| chr4_129928333_129928910 | 0.03 |
Spocd1 |
SPOC domain containing 1 |
628 |
0.61 |
| chr17_67828303_67828454 | 0.03 |
Lama1 |
laminin, alpha 1 |
15972 |
0.2 |
| chr4_40852424_40852797 | 0.03 |
B4galt1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
1395 |
0.24 |
| chr8_70891263_70891847 | 0.03 |
Slc5a5 |
solute carrier family 5 (sodium iodide symporter), member 5 |
1202 |
0.23 |
| chr3_86550029_86551252 | 0.03 |
Mab21l2 |
mab-21-like 2 |
2011 |
0.32 |
| chr8_83575796_83577746 | 0.03 |
D830024N08Rik |
RIKEN cDNA D830024N08 gene |
727 |
0.44 |
| chr10_80380857_80382709 | 0.03 |
Mex3d |
mex3 RNA binding family member D |
204 |
0.7 |
| chr15_103365583_103366152 | 0.03 |
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
881 |
0.44 |
| chr11_82841815_82841966 | 0.03 |
Rffl |
ring finger and FYVE like domain containing protein |
4202 |
0.14 |
| chr2_27677433_27678576 | 0.03 |
Rxra |
retinoid X receptor alpha |
742 |
0.75 |
| chr10_127520039_127520190 | 0.03 |
Shmt2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
1334 |
0.26 |
| chr19_5756395_5757941 | 0.03 |
Ltbp3 |
latent transforming growth factor beta binding protein 3 |
198 |
0.83 |
| chr6_86547683_86548400 | 0.02 |
1600020E01Rik |
RIKEN cDNA 1600020E01 gene |
60 |
0.95 |
| chr5_136965439_136966847 | 0.02 |
Cldn15 |
claudin 15 |
473 |
0.63 |
| chr3_96384099_96384480 | 0.02 |
Rnu1b2 |
U1b2 small nuclear RNA |
10268 |
0.06 |
| chr1_191574277_191575491 | 0.02 |
Dtl |
denticleless E3 ubiquitin protein ligase |
627 |
0.46 |
| chr1_163361816_163362288 | 0.02 |
Gm24940 |
predicted gene, 24940 |
40231 |
0.12 |
| chr3_146117173_146118635 | 0.02 |
Mcoln3 |
mucolipin 3 |
454 |
0.73 |
| chr4_115938793_115939428 | 0.02 |
Dmbx1 |
diencephalon/mesencephalon homeobox 1 |
816 |
0.55 |
| chr17_35045614_35047458 | 0.02 |
Msh5 |
mutS homolog 5 |
6 |
0.9 |
| chr19_12575704_12576006 | 0.02 |
Fam111a |
family with sequence similarity 111, member A |
2332 |
0.16 |
| chr13_35741218_35741875 | 0.02 |
Cdyl |
chromodomain protein, Y chromosome-like |
233 |
0.93 |
| chr12_75616485_75616662 | 0.02 |
AC124742.1 |
ribosomal protein, large P2 (Rplp2), retrogene |
14745 |
0.17 |
| chr6_88198649_88199998 | 0.02 |
Gata2 |
GATA binding protein 2 |
989 |
0.43 |
| chr12_84191503_84191654 | 0.02 |
Elmsan1 |
ELM2 and Myb/SANT-like domain containing 1 |
2437 |
0.16 |
| chr11_88860478_88860629 | 0.02 |
Akap1 |
A kinase (PRKA) anchor protein 1 |
3164 |
0.2 |
| chr1_193382739_193383431 | 0.02 |
Camk1g |
calcium/calmodulin-dependent protein kinase I gamma |
12787 |
0.15 |
| chr13_54737787_54738662 | 0.02 |
Gprin1 |
G protein-regulated inducer of neurite outgrowth 1 |
11445 |
0.12 |
| chr5_33433256_33435017 | 0.02 |
Nkx1-1 |
NK1 homeobox 1 |
160 |
0.94 |
| chr4_3230864_3231239 | 0.02 |
AI838599 |
expressed sequence AI838599 |
11004 |
0.17 |
| chr1_58029648_58030887 | 0.02 |
Aox1 |
aldehyde oxidase 1 |
303 |
0.89 |
| chr7_44747810_44748395 | 0.02 |
Zfp473 |
zinc finger protein 473 |
53 |
0.79 |
| chr18_76034867_76035898 | 0.02 |
1700003O11Rik |
RIKEN cDNA 1700003O11 gene |
19705 |
0.19 |
| chr5_107196279_107196430 | 0.02 |
Tgfbr3 |
transforming growth factor, beta receptor III |
3987 |
0.19 |
| chr11_69047580_69047908 | 0.02 |
Aurkb |
aurora kinase B |
233 |
0.81 |
| chr11_101784179_101785383 | 0.02 |
Etv4 |
ets variant 4 |
67 |
0.97 |
| chr5_140815321_140815472 | 0.02 |
Gna12 |
guanine nucleotide binding protein, alpha 12 |
14761 |
0.2 |
| chr9_44733806_44735185 | 0.02 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
703 |
0.43 |
| chr9_21851806_21852608 | 0.02 |
Dock6 |
dedicator of cytokinesis 6 |
409 |
0.74 |
| chr4_48407474_48407947 | 0.02 |
Invs |
inversin |
25522 |
0.19 |
| chr10_19779040_19779708 | 0.02 |
Gm25256 |
predicted gene, 25256 |
3480 |
0.25 |
| chr17_71001223_71003092 | 0.02 |
Myl12a |
myosin, light chain 12A, regulatory, non-sarcomeric |
3 |
0.49 |
| chr4_135913823_135913974 | 0.02 |
Cnr2 |
cannabinoid receptor 2 (macrophage) |
2080 |
0.18 |
| chr7_126492285_126492979 | 0.02 |
Atxn2l |
ataxin 2-like |
744 |
0.45 |
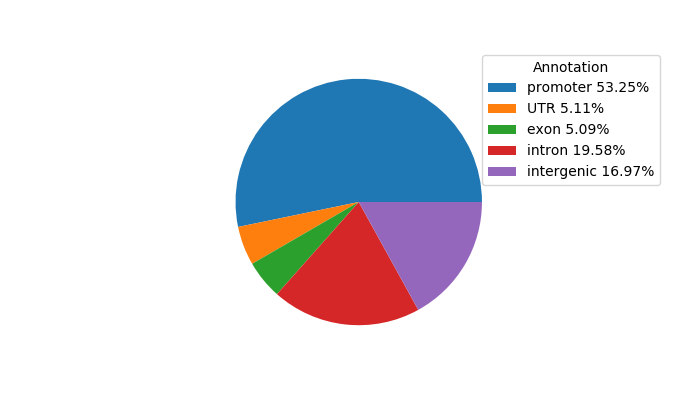
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |