Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
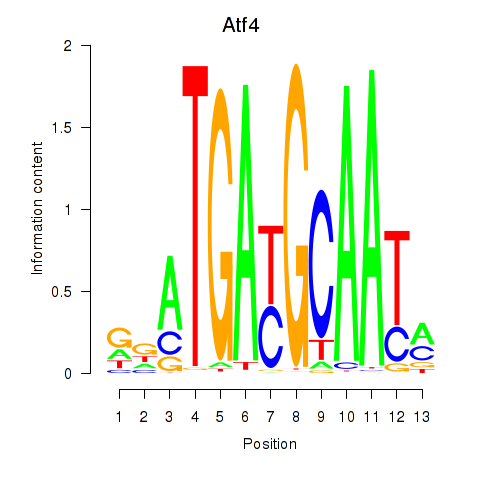
Results for Atf4
Z-value: 0.59
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSMUSG00000042406.7 | activating transcription factor 4 |
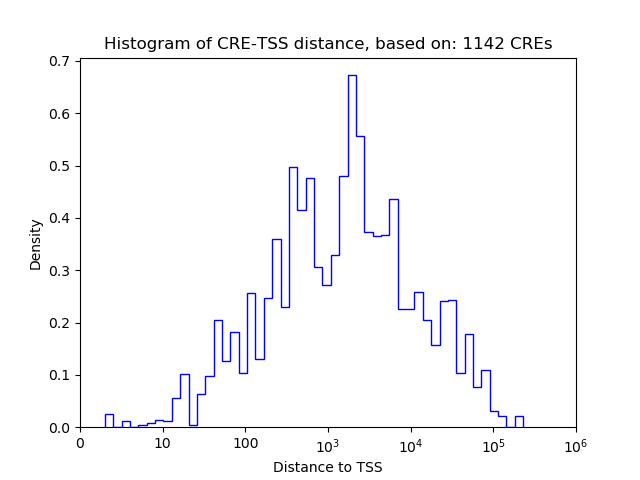
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_80254231_80254977 | Atf4 | 580 | 0.599683 | 0.37 | 3.6e-03 | Click! |
| chr15_80257273_80257434 | Atf4 | 1830 | 0.207522 | 0.25 | 5.8e-02 | Click! |
| chr15_80257030_80257248 | Atf4 | 1616 | 0.232875 | 0.23 | 7.3e-02 | Click! |
| chr15_80256684_80256930 | Atf4 | 1284 | 0.292459 | 0.19 | 1.4e-01 | Click! |
| chr15_80257680_80258950 | Atf4 | 2792 | 0.150338 | 0.17 | 1.9e-01 | Click! |
Activity of the Atf4 motif across conditions
Conditions sorted by the z-value of the Atf4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
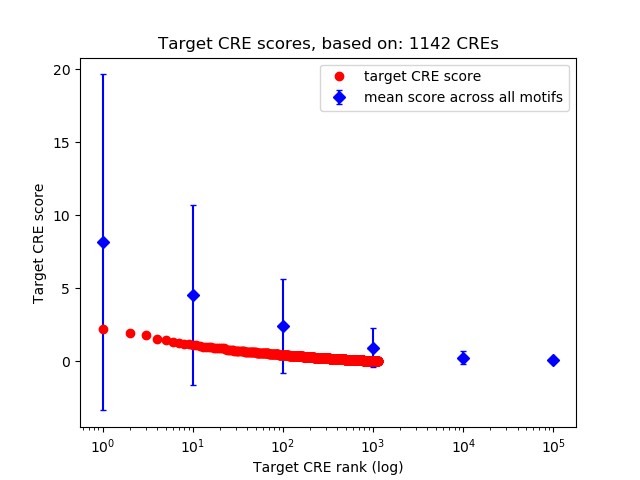
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_32123581_32123732 | 2.20 |
5430401H09Rik |
RIKEN cDNA 5430401H09 gene |
46 |
0.98 |
| chr14_70625458_70627688 | 1.97 |
Dmtn |
dematin actin binding protein |
418 |
0.75 |
| chr11_6598568_6599983 | 1.79 |
Nacad |
NAC alpha domain containing |
117 |
0.92 |
| chr8_121897389_121899362 | 1.50 |
Slc7a5 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
1873 |
0.2 |
| chr2_146098449_146098737 | 1.49 |
Cfap61 |
cilia and flagella associated protein 61 |
51342 |
0.15 |
| chr17_36866497_36867750 | 1.34 |
Trim15 |
tripartite motif-containing 15 |
64 |
0.93 |
| chr11_11687647_11688276 | 1.23 |
Ikzf1 |
IKAROS family zinc finger 1 |
1516 |
0.33 |
| chr1_153660970_153662018 | 1.22 |
Rgs8 |
regulator of G-protein signaling 8 |
413 |
0.8 |
| chr7_61311164_61311772 | 1.16 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
245 |
0.96 |
| chr7_61159018_61159292 | 1.11 |
Gm38451 |
predicted gene, 38451 |
28471 |
0.24 |
| chr12_111442182_111444685 | 1.10 |
Tnfaip2 |
tumor necrosis factor, alpha-induced protein 2 |
771 |
0.51 |
| chr19_43985320_43986721 | 1.04 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
536 |
0.73 |
| chr7_61221429_61221953 | 1.01 |
A230006K03Rik |
RIKEN cDNA A230006K03 gene |
90022 |
0.08 |
| chr2_172393314_172394463 | 0.99 |
Cass4 |
Cas scaffolding protein family member 4 |
12 |
0.97 |
| chr1_79437543_79437779 | 0.98 |
Scg2 |
secretogranin II |
2381 |
0.37 |
| chr7_75853296_75853470 | 0.97 |
Klhl25 |
kelch-like 25 |
4942 |
0.25 |
| chr2_119350553_119351941 | 0.94 |
Chac1 |
ChaC, cation transport regulator 1 |
18 |
0.97 |
| chr1_167383214_167383424 | 0.92 |
Mgst3 |
microsomal glutathione S-transferase 3 |
10522 |
0.14 |
| chr10_127511678_127514192 | 0.91 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
2032 |
0.18 |
| chr7_48791267_48791418 | 0.90 |
Zdhhc13 |
zinc finger, DHHC domain containing 13 |
2310 |
0.24 |
| chr8_85380167_85381092 | 0.89 |
Mylk3 |
myosin light chain kinase 3 |
349 |
0.83 |
| chr15_84324521_84325304 | 0.89 |
Parvg |
parvin, gamma |
117 |
0.94 |
| chr2_129309240_129309540 | 0.83 |
Il1a |
interleukin 1 alpha |
582 |
0.59 |
| chr7_16784339_16784989 | 0.79 |
Slc1a5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
74 |
0.95 |
| chr3_108444057_108444211 | 0.77 |
Sars |
seryl-aminoacyl-tRNA synthetase |
1009 |
0.32 |
| chr17_32117457_32118702 | 0.77 |
Gm17276 |
predicted gene, 17276 |
4332 |
0.15 |
| chr12_103388537_103389963 | 0.76 |
Otub2 |
OTU domain, ubiquitin aldehyde binding 2 |
265 |
0.84 |
| chr17_31033726_31034632 | 0.75 |
Abcg1 |
ATP binding cassette subfamily G member 1 |
23496 |
0.11 |
| chr11_95778707_95778988 | 0.74 |
Polr2k-ps |
polymerase (RNA) II (DNA directed) polypeptide K, pseudogene |
17356 |
0.11 |
| chr8_70630745_70631808 | 0.74 |
Gdf15 |
growth differentiation factor 15 |
819 |
0.39 |
| chr4_117835381_117836667 | 0.72 |
Slc6a9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
586 |
0.62 |
| chr6_45062685_45062917 | 0.70 |
Cntnap2 |
contactin associated protein-like 2 |
2740 |
0.24 |
| chr11_24092672_24093146 | 0.70 |
Bcl11a |
B cell CLL/lymphoma 11A (zinc finger protein) |
12239 |
0.14 |
| chr15_102150305_102151233 | 0.69 |
Soat2 |
sterol O-acyltransferase 2 |
243 |
0.86 |
| chr17_32091482_32092224 | 0.68 |
Pdxk-ps |
pyridoxal (pyridoxine, vitamin B6) kinase, pseudogene |
430 |
0.78 |
| chr9_96733821_96734335 | 0.68 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
1308 |
0.4 |
| chr4_115061778_115062015 | 0.67 |
Tal1 |
T cell acute lymphocytic leukemia 1 |
2388 |
0.24 |
| chr8_54956899_54957247 | 0.66 |
Gpm6a |
glycoprotein m6a |
2230 |
0.24 |
| chr2_69341375_69341619 | 0.66 |
Abcb11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
1103 |
0.5 |
| chr2_26359770_26360886 | 0.66 |
Card9 |
caspase recruitment domain family, member 9 |
581 |
0.52 |
| chr1_195129782_195130181 | 0.65 |
Cr1l |
complement component (3b/4b) receptor 1-like |
519 |
0.72 |
| chr3_83010068_83011013 | 0.65 |
Gm30097 |
predicted gene, 30097 |
2052 |
0.23 |
| chr13_4233789_4234486 | 0.65 |
Akr1c19 |
aldo-keto reductase family 1, member C19 |
397 |
0.81 |
| chr7_122672758_122673250 | 0.65 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
1586 |
0.39 |
| chr3_57426842_57427133 | 0.64 |
Tm4sf4 |
transmembrane 4 superfamily member 4 |
1673 |
0.42 |
| chr2_118554047_118554750 | 0.64 |
Bmf |
BCL2 modifying factor |
4711 |
0.18 |
| chr13_101691106_101693172 | 0.63 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
204 |
0.95 |
| chr4_108544443_108544594 | 0.63 |
Tut4 |
terminal uridylyl transferase 4 |
2100 |
0.17 |
| chr5_124192871_124193356 | 0.62 |
Pitpnm2os2 |
phosphatidylinositol transfer protein, membrane-associated 2, opposite strand 2 |
1794 |
0.23 |
| chr1_9745419_9746231 | 0.61 |
1700034P13Rik |
RIKEN cDNA 1700034P13 gene |
1823 |
0.26 |
| chr3_95950652_95951560 | 0.59 |
Anp32e |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
6902 |
0.1 |
| chr18_56978049_56978296 | 0.59 |
C330018D20Rik |
RIKEN cDNA C330018D20 gene |
2804 |
0.3 |
| chr4_19704870_19705084 | 0.59 |
Wwp1 |
WW domain containing E3 ubiquitin protein ligase 1 |
4016 |
0.25 |
| chr14_60637402_60638329 | 0.58 |
Spata13 |
spermatogenesis associated 13 |
3110 |
0.3 |
| chr12_78904719_78905643 | 0.58 |
Plek2 |
pleckstrin 2 |
1783 |
0.34 |
| chr7_78913499_78914279 | 0.58 |
Isg20 |
interferon-stimulated protein |
92 |
0.95 |
| chr11_11962671_11962822 | 0.58 |
Grb10 |
growth factor receptor bound protein 10 |
6722 |
0.22 |
| chr4_141073557_141074133 | 0.57 |
Necap2 |
NECAP endocytosis associated 2 |
140 |
0.93 |
| chr6_69500271_69500422 | 0.57 |
Gm47304 |
predicted gene, 47304 |
16135 |
0.12 |
| chr11_108607827_108608389 | 0.57 |
Cep112 |
centrosomal protein 112 |
2881 |
0.32 |
| chr4_132078510_132078882 | 0.57 |
Epb41 |
erythrocyte membrane protein band 4.1 |
3375 |
0.14 |
| chr9_44339477_44339803 | 0.57 |
Hmbs |
hydroxymethylbilane synthase |
76 |
0.9 |
| chr3_59130395_59131019 | 0.57 |
P2ry14 |
purinergic receptor P2Y, G-protein coupled, 14 |
55 |
0.97 |
| chr6_149099657_149099981 | 0.56 |
Dennd5b |
DENN/MADD domain containing 5B |
1719 |
0.23 |
| chr10_11151671_11152032 | 0.55 |
Shprh |
SNF2 histone linker PHD RING helicase |
2327 |
0.34 |
| chr14_16847192_16847623 | 0.55 |
Rarb |
retinoic acid receptor, beta |
28251 |
0.23 |
| chr6_144779851_144780433 | 0.54 |
Sox5 |
SRY (sex determining region Y)-box 5 |
1777 |
0.35 |
| chr7_109165724_109166870 | 0.54 |
Lmo1 |
LIM domain only 1 |
4215 |
0.21 |
| chrX_85616079_85616575 | 0.53 |
Gm44378 |
predicted gene, 44378 |
27350 |
0.18 |
| chr15_89322584_89323784 | 0.53 |
Adm2 |
adrenomedullin 2 |
464 |
0.63 |
| chr7_126624812_126625701 | 0.53 |
Nupr1 |
nuclear protein transcription regulator 1 |
88 |
0.92 |
| chr13_34341882_34342941 | 0.53 |
Slc22a23 |
solute carrier family 22, member 23 |
1792 |
0.34 |
| chr5_148398928_148400150 | 0.53 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
252 |
0.94 |
| chr13_91372302_91372553 | 0.52 |
Gm29540 |
predicted gene 29540 |
1867 |
0.37 |
| chr14_27430281_27430617 | 0.52 |
Tasor |
transcription activation suppressor |
1615 |
0.38 |
| chr7_115859202_115859391 | 0.51 |
Sox6 |
SRY (sex determining region Y)-box 6 |
556 |
0.85 |
| chr13_19822928_19823079 | 0.51 |
Gpr141 |
G protein-coupled receptor 141 |
1224 |
0.45 |
| chr9_71156685_71156836 | 0.51 |
Aqp9 |
aquaporin 9 |
5873 |
0.19 |
| chr11_17007546_17008653 | 0.50 |
Plek |
pleckstrin |
603 |
0.7 |
| chr3_89997986_89998664 | 0.50 |
Hax1 |
HCLS1 associated X-1 |
37 |
0.85 |
| chr6_126735053_126735582 | 0.49 |
Kcna6 |
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
1494 |
0.38 |
| chr8_94387801_94388063 | 0.49 |
Herpud1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
333 |
0.71 |
| chr8_35882704_35883048 | 0.48 |
5430403N17Rik |
RIKEN cDNA 5430403N17 gene |
81 |
0.97 |
| chr8_121850755_121851753 | 0.48 |
Gm26812 |
predicted gene, 26812 |
629 |
0.55 |
| chr13_35755922_35756175 | 0.48 |
Cdyl |
chromodomain protein, Y chromosome-like |
6764 |
0.19 |
| chr12_116281089_116281240 | 0.47 |
Esyt2 |
extended synaptotagmin-like protein 2 |
32 |
0.94 |
| chr6_134927945_134928524 | 0.47 |
Lockd |
lncRNA downstream of Cdkn1b |
858 |
0.42 |
| chr9_77435194_77435444 | 0.47 |
Lrrc1 |
leucine rich repeat containing 1 |
6876 |
0.2 |
| chr14_54417149_54418129 | 0.46 |
Slc7a7 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
40 |
0.94 |
| chr18_34778689_34779380 | 0.46 |
Kdm3b |
KDM3B lysine (K)-specific demethylase 3B |
1929 |
0.22 |
| chr2_65620767_65621991 | 0.46 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
568 |
0.82 |
| chr7_135588799_135589009 | 0.46 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
4484 |
0.21 |
| chr19_3762115_3762266 | 0.46 |
Kmt5b |
lysine methyltransferase 5B |
5231 |
0.11 |
| chr2_4572310_4572962 | 0.46 |
Frmd4a |
FERM domain containing 4A |
6460 |
0.21 |
| chr6_55038035_55039062 | 0.46 |
Gars |
glycyl-tRNA synthetase |
541 |
0.77 |
| chr8_113551629_113551780 | 0.46 |
Gm24291 |
predicted gene, 24291 |
23033 |
0.19 |
| chr6_99191228_99191426 | 0.45 |
Foxp1 |
forkhead box P1 |
28309 |
0.24 |
| chr14_60639788_60640342 | 0.44 |
Spata13 |
spermatogenesis associated 13 |
5310 |
0.25 |
| chr13_45904596_45904786 | 0.44 |
4930453C13Rik |
RIKEN cDNA 4930453C13 gene |
31349 |
0.17 |
| chr5_110836247_110836790 | 0.44 |
Hscb |
HscB iron-sulfur cluster co-chaperone |
198 |
0.9 |
| chr12_72763251_72763786 | 0.44 |
Ppm1a |
protein phosphatase 1A, magnesium dependent, alpha isoform |
2203 |
0.3 |
| chr17_35000871_35002234 | 0.44 |
Vars |
valyl-tRNA synthetase |
235 |
0.74 |
| chr12_107994446_107995262 | 0.44 |
Bcl11b |
B cell leukemia/lymphoma 11B |
8560 |
0.29 |
| chr11_100320488_100321246 | 0.43 |
Eif1 |
eukaryotic translation initiation factor 1 |
17 |
0.94 |
| chr4_120856851_120857002 | 0.43 |
Rims3 |
regulating synaptic membrane exocytosis 3 |
2107 |
0.21 |
| chr7_111179382_111180585 | 0.43 |
1700012D14Rik |
RIKEN cDNA 1700012D14 gene |
57299 |
0.11 |
| chr17_88066306_88066664 | 0.43 |
Fbxo11 |
F-box protein 11 |
1194 |
0.51 |
| chr12_73779438_73779861 | 0.42 |
Gm8075 |
predicted gene 8075 |
13834 |
0.2 |
| chr18_65802028_65802182 | 0.42 |
Sec11c |
SEC11 homolog C, signal peptidase complex subunit |
803 |
0.51 |
| chr15_86181872_86183211 | 0.42 |
Gm15569 |
predicted gene 15569 |
3187 |
0.2 |
| chr6_67034247_67034846 | 0.41 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
2053 |
0.19 |
| chr7_126776991_126777669 | 0.41 |
Ypel3 |
yippee like 3 |
117 |
0.78 |
| chr1_37433025_37433308 | 0.41 |
Unc50 |
unc-50 homolog |
1822 |
0.27 |
| chr15_100738581_100738785 | 0.41 |
I730030J21Rik |
RIKEN cDNA I730030J21 gene |
5946 |
0.13 |
| chr2_91268902_91269542 | 0.40 |
Arfgap2 |
ADP-ribosylation factor GTPase activating protein 2 |
120 |
0.94 |
| chr4_140699463_140700390 | 0.40 |
Rcc2 |
regulator of chromosome condensation 2 |
615 |
0.61 |
| chr11_20200566_20201086 | 0.40 |
Rab1a |
RAB1A, member RAS oncogene family |
606 |
0.7 |
| chr7_61030064_61030449 | 0.40 |
Gm44643 |
predicted gene 44643 |
68871 |
0.12 |
| chr11_88614518_88614971 | 0.40 |
Msi2 |
musashi RNA-binding protein 2 |
24597 |
0.21 |
| chr11_83262776_83263197 | 0.40 |
Gm11427 |
predicted gene 11427 |
7841 |
0.09 |
| chr18_69502058_69502470 | 0.40 |
Tcf4 |
transcription factor 4 |
1306 |
0.56 |
| chr7_19022230_19023942 | 0.40 |
Foxa3 |
forkhead box A3 |
452 |
0.57 |
| chr8_25368281_25369181 | 0.39 |
Gm39147 |
predicted gene, 39147 |
30289 |
0.13 |
| chr7_102444287_102444743 | 0.39 |
Rrm1 |
ribonucleotide reductase M1 |
2472 |
0.15 |
| chr6_31125380_31126701 | 0.38 |
5330406M23Rik |
RIKEN cDNA 5330406M23 gene |
15120 |
0.11 |
| chr4_118078652_118079394 | 0.38 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
47141 |
0.11 |
| chr4_89485854_89486155 | 0.38 |
Gm12608 |
predicted gene 12608 |
41360 |
0.16 |
| chr19_29203241_29203392 | 0.38 |
Gm5518 |
predicted gene 5518 |
43198 |
0.11 |
| chr6_120956618_120956869 | 0.38 |
Mical3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
980 |
0.41 |
| chr4_43957708_43958633 | 0.38 |
Glipr2 |
GLI pathogenesis-related 2 |
478 |
0.74 |
| chr11_24129305_24129928 | 0.38 |
Bcl11a |
B cell CLL/lymphoma 11A (zinc finger protein) |
12763 |
0.15 |
| chr2_167043825_167044361 | 0.37 |
Znfx1 |
zinc finger, NFX1-type containing 1 |
598 |
0.55 |
| chr7_35055557_35056579 | 0.37 |
Gm45095 |
predicted gene 45095 |
41 |
0.53 |
| chr10_39369812_39370687 | 0.37 |
Fyn |
Fyn proto-oncogene |
446 |
0.85 |
| chr12_84204289_84204451 | 0.37 |
Gm31513 |
predicted gene, 31513 |
8401 |
0.11 |
| chr5_21056286_21056819 | 0.37 |
Ptpn12 |
protein tyrosine phosphatase, non-receptor type 12 |
641 |
0.68 |
| chr9_16437830_16438127 | 0.37 |
Gm48726 |
predicted gene, 48726 |
2583 |
0.4 |
| chr10_80331489_80332188 | 0.37 |
Reep6 |
receptor accessory protein 6 |
1636 |
0.14 |
| chr15_81802115_81803592 | 0.37 |
Tef |
thyrotroph embryonic factor |
27 |
0.75 |
| chr5_118482858_118484042 | 0.37 |
Gm15754 |
predicted gene 15754 |
3517 |
0.25 |
| chr13_100512536_100512862 | 0.37 |
Gtf2h2 |
general transcription factor II H, polypeptide 2 |
20120 |
0.12 |
| chr3_97585683_97585944 | 0.36 |
Chd1l |
chromodomain helicase DNA binding protein 1-like |
18701 |
0.15 |
| chr7_92887260_92887411 | 0.36 |
Prcp |
prolylcarboxypeptidase (angiotensinase C) |
12035 |
0.15 |
| chr8_123856422_123856573 | 0.36 |
Gm4342 |
predicted gene 4342 |
2668 |
0.13 |
| chr10_58370178_58370662 | 0.36 |
Lims1 |
LIM and senescent cell antigen-like domains 1 |
1034 |
0.53 |
| chr13_73476363_73476514 | 0.36 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
4687 |
0.25 |
| chr11_83283019_83283399 | 0.36 |
Slfn14 |
schlafen 14 |
3517 |
0.11 |
| chr18_67241729_67242343 | 0.36 |
Mppe1 |
metallophosphoesterase 1 |
3241 |
0.19 |
| chr19_10305676_10305827 | 0.36 |
Dagla |
diacylglycerol lipase, alpha |
874 |
0.53 |
| chr7_61614761_61615352 | 0.36 |
B230209E15Rik |
RIKEN cDNA B230209E15 gene |
271 |
0.89 |
| chr2_72324313_72324464 | 0.36 |
Map3k20 |
mitogen-activated protein kinase kinase kinase 20 |
26487 |
0.16 |
| chr3_91092756_91092907 | 0.36 |
S100a7l2 |
S100 calcium binding protein A7 like 2 |
2028 |
0.34 |
| chr5_142650560_142650759 | 0.36 |
Wipi2 |
WD repeat domain, phosphoinositide interacting 2 |
11855 |
0.15 |
| chr19_53325039_53326410 | 0.35 |
Gm30541 |
predicted gene, 30541 |
1 |
0.97 |
| chr1_189341351_189342108 | 0.35 |
Kcnk2 |
potassium channel, subfamily K, member 2 |
1626 |
0.32 |
| chr3_153721362_153721840 | 0.35 |
St6galnac3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
605 |
0.69 |
| chr3_73051639_73051897 | 0.35 |
Slitrk3 |
SLIT and NTRK-like family, member 3 |
5175 |
0.2 |
| chr15_102789128_102789616 | 0.35 |
Gm49473 |
predicted gene, 49473 |
5999 |
0.16 |
| chr14_45655772_45656240 | 0.35 |
Ddhd1 |
DDHD domain containing 1 |
1354 |
0.28 |
| chr4_123525767_123526146 | 0.35 |
Macf1 |
microtubule-actin crosslinking factor 1 |
1503 |
0.44 |
| chr7_6435207_6435948 | 0.34 |
Olfr1344 |
olfactory receptor 1344 |
3111 |
0.1 |
| chr10_41209464_41210153 | 0.34 |
Gm25526 |
predicted gene, 25526 |
6237 |
0.22 |
| chr4_115839985_115840231 | 0.34 |
Mknk1 |
MAP kinase-interacting serine/threonine kinase 1 |
848 |
0.49 |
| chr9_90249400_90250047 | 0.33 |
Tbc1d2b |
TBC1 domain family, member 2B |
6204 |
0.18 |
| chr9_87021571_87022041 | 0.33 |
Cyb5r4 |
cytochrome b5 reductase 4 |
208 |
0.93 |
| chr6_121244244_121244395 | 0.33 |
Usp18 |
ubiquitin specific peptidase 18 |
1587 |
0.28 |
| chr9_70543545_70543696 | 0.32 |
Sltm |
SAFB-like, transcription modulator |
722 |
0.58 |
| chr12_108893486_108894172 | 0.32 |
Wdr25 |
WD repeat domain 25 |
198 |
0.57 |
| chr7_4121960_4122900 | 0.32 |
Ttyh1 |
tweety family member 1 |
281 |
0.78 |
| chr16_91043345_91044458 | 0.32 |
Paxbp1 |
PAX3 and PAX7 binding protein 1 |
263 |
0.76 |
| chr17_25961783_25962611 | 0.31 |
Capn15 |
calpain 15 |
1563 |
0.17 |
| chr2_165887152_165887879 | 0.31 |
Zmynd8 |
zinc finger, MYND-type containing 8 |
2641 |
0.19 |
| chr8_54954519_54955779 | 0.31 |
Gpm6a |
glycoprotein m6a |
306 |
0.88 |
| chr6_22025534_22026744 | 0.31 |
Cped1 |
cadherin-like and PC-esterase domain containing 1 |
196 |
0.96 |
| chr1_58970537_58971332 | 0.31 |
Trak2 |
trafficking protein, kinesin binding 2 |
2495 |
0.2 |
| chr5_96912273_96912506 | 0.31 |
Gm43147 |
predicted gene 43147 |
852 |
0.41 |
| chr17_47010961_47011192 | 0.31 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
520 |
0.76 |
| chr2_26485135_26488628 | 0.30 |
Notch1 |
notch 1 |
16383 |
0.09 |
| chr7_101684127_101684485 | 0.30 |
Clpb |
ClpB caseinolytic peptidase B |
1109 |
0.51 |
| chr5_112244110_112244261 | 0.30 |
Cryba4 |
crystallin, beta A4 |
6977 |
0.12 |
| chrX_134620328_134620674 | 0.30 |
Gm7855 |
predicted gene 7855 |
5243 |
0.12 |
| chrX_23281419_23281815 | 0.30 |
Klhl13 |
kelch-like 13 |
3212 |
0.36 |
| chr6_3988317_3988783 | 0.30 |
Tfpi2 |
tissue factor pathway inhibitor 2 |
308 |
0.88 |
| chr12_5371578_5371994 | 0.30 |
Klhl29 |
kelch-like 29 |
3896 |
0.26 |
| chrX_66657509_66657976 | 0.30 |
Slitrk2 |
SLIT and NTRK-like family, member 2 |
4735 |
0.2 |
| chr19_61153275_61153617 | 0.29 |
Gm22365 |
predicted gene, 22365 |
11035 |
0.12 |
| chr8_122636390_122636583 | 0.29 |
Cbfa2t3 |
CBFA2/RUNX1 translocation partner 3 |
9980 |
0.09 |
| chr3_84313220_84313371 | 0.29 |
Trim2 |
tripartite motif-containing 2 |
6418 |
0.27 |
| chr13_114531946_114532233 | 0.29 |
Gm25950 |
predicted gene, 25950 |
48406 |
0.11 |
| chr1_66386919_66387899 | 0.29 |
Map2 |
microtubule-associated protein 2 |
398 |
0.87 |
| chr5_81196201_81196352 | 0.29 |
Gm43594 |
predicted gene 43594 |
52973 |
0.17 |
| chr19_44106802_44107431 | 0.29 |
Chuk |
conserved helix-loop-helix ubiquitous kinase |
166 |
0.92 |
| chr17_53569556_53570941 | 0.28 |
Kat2b |
K(lysine) acetyltransferase 2B |
3277 |
0.2 |
| chr7_109602204_109602761 | 0.28 |
Denn2b |
DENN domain containing 2B |
183 |
0.94 |
| chr7_121709089_121709240 | 0.28 |
Usp31 |
ubiquitin specific peptidase 31 |
1911 |
0.27 |
| chr15_103252820_103253410 | 0.28 |
Nfe2 |
nuclear factor, erythroid derived 2 |
37 |
0.95 |
| chr3_88831100_88832914 | 0.28 |
1500004A13Rik |
RIKEN cDNA 1500004A13 gene |
361 |
0.77 |
| chr1_178187713_178188786 | 0.28 |
Desi2 |
desumoylating isopeptidase 2 |
402 |
0.86 |
| chr3_103172753_103173085 | 0.28 |
Bcas2 |
breast carcinoma amplified sequence 2 |
1134 |
0.38 |
| chr11_46077651_46077829 | 0.28 |
Adam19 |
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
21682 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 1.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.5 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.8 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 | 0.8 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.5 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.6 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.6 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 0.2 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.4 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.1 | 0.6 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 | 0.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.3 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.6 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 0.2 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 0.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.9 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0060051 | negative regulation of protein glycosylation(GO:0060051) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.4 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0042523 | positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.0 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:1901741 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.0 | GO:0009130 | UMP biosynthetic process(GO:0006222) pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.3 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005818 | aster(GO:0005818) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 0.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.2 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:0043176 | amine binding(GO:0043176) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.6 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0034534 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0004337 | geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |