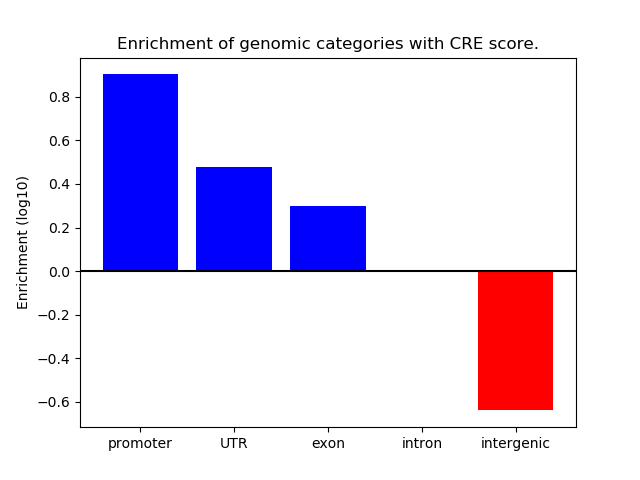
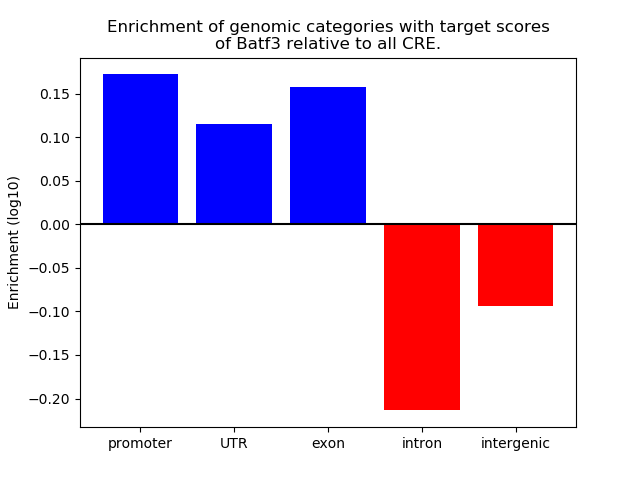
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
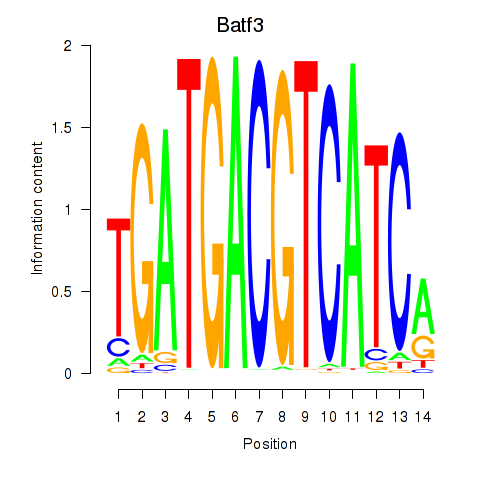
Results for Batf3
Z-value: 0.50
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.4 | basic leucine zipper transcription factor, ATF-like 3 |
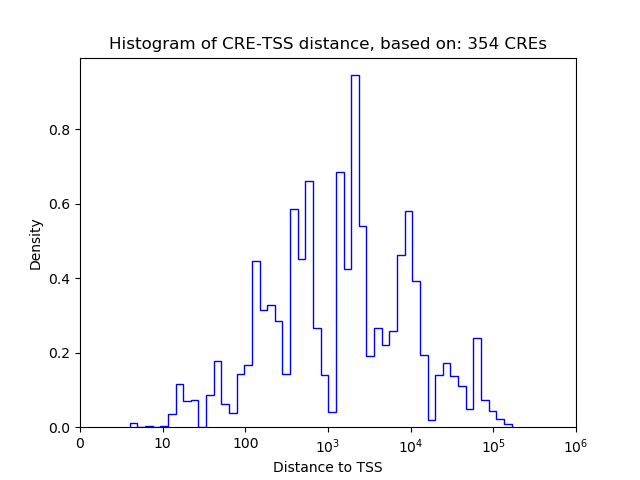
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_191097653_191098840 | Batf3 | 79 | 0.950610 | -0.36 | 4.8e-03 | Click! |
| chr1_191097128_191097279 | Batf3 | 644 | 0.578474 | -0.15 | 2.7e-01 | Click! |
| chr1_191111232_191111383 | Batf3 | 10830 | 0.113981 | -0.12 | 3.7e-01 | Click! |
Activity of the Batf3 motif across conditions
Conditions sorted by the z-value of the Batf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
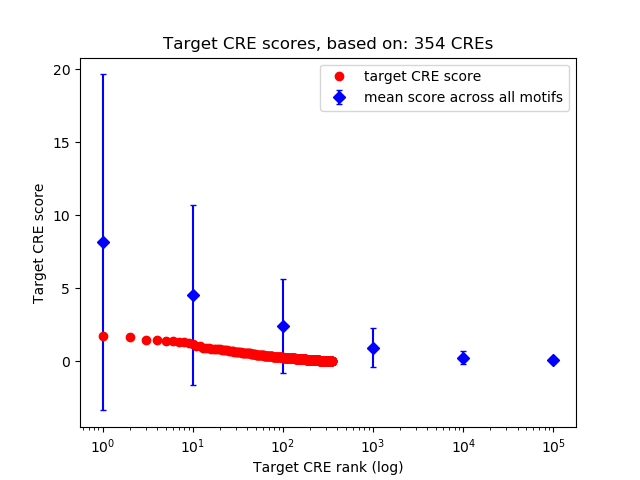
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_116572624_116573264 | 1.74 |
Ube2o |
ubiquitin-conjugating enzyme E2O |
8503 |
0.1 |
| chr4_142017816_142018715 | 1.69 |
4930455G09Rik |
RIKEN cDNA 4930455G09 gene |
367 |
0.8 |
| chr4_46040988_46042013 | 1.45 |
Tmod1 |
tropomodulin 1 |
2291 |
0.3 |
| chr17_34998907_34999597 | 1.43 |
Vars |
valyl-tRNA synthetase |
1735 |
0.11 |
| chr19_5686209_5687218 | 1.42 |
Pcnx3 |
pecanex homolog 3 |
1436 |
0.16 |
| chr6_57822563_57823744 | 1.38 |
Vopp1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
1487 |
0.3 |
| chr6_31651754_31651924 | 1.35 |
Gm43154 |
predicted gene 43154 |
797 |
0.64 |
| chr15_58575323_58575591 | 1.29 |
Fer1l6 |
fer-1-like 6 (C. elegans) |
63048 |
0.12 |
| chr15_27871217_27871844 | 1.26 |
Gm20555 |
predicted gene, 20555 |
4299 |
0.22 |
| chr8_60953813_60955404 | 1.20 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
140 |
0.95 |
| chr7_128414548_128415092 | 1.07 |
Gm15503 |
predicted gene 15503 |
2479 |
0.18 |
| chr17_53596477_53596632 | 1.05 |
Gm6919 |
predicted gene 6919 |
2109 |
0.26 |
| chr8_122306253_122307213 | 0.93 |
Zfpm1 |
zinc finger protein, multitype 1 |
587 |
0.68 |
| chr7_141146140_141146436 | 0.90 |
Ptdss2 |
phosphatidylserine synthase 2 |
8335 |
0.08 |
| chr3_127468377_127468752 | 0.88 |
Gm44494 |
predicted gene, 44494 |
12556 |
0.1 |
| chr8_70500810_70501374 | 0.87 |
Crlf1 |
cytokine receptor-like factor 1 |
41 |
0.94 |
| chr6_86472299_86472450 | 0.86 |
C87436 |
expressed sequence C87436 |
8334 |
0.08 |
| chr4_155893812_155895517 | 0.83 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2352 |
0.11 |
| chr16_92340779_92341614 | 0.83 |
Smim34 |
small integral membrane protein 34 |
11942 |
0.11 |
| chr2_49730442_49730664 | 0.83 |
Kif5c |
kinesin family member 5C |
2058 |
0.38 |
| chr17_28013120_28014046 | 0.79 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
6238 |
0.13 |
| chr11_115511512_115512231 | 0.77 |
Jpt1 |
Jupiter microtubule associated homolog 1 |
2245 |
0.14 |
| chr7_142088584_142090929 | 0.77 |
Dusp8 |
dual specificity phosphatase 8 |
5516 |
0.09 |
| chr1_75214392_75214921 | 0.74 |
Stk16 |
serine/threonine kinase 16 |
1595 |
0.14 |
| chr10_80140825_80141682 | 0.72 |
Atp5d |
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
204 |
0.67 |
| chr13_21935384_21936058 | 0.71 |
Zfp184 |
zinc finger protein 184 (Kruppel-like) |
9373 |
0.06 |
| chr12_109543555_109544268 | 0.71 |
Meg3 |
maternally expressed 3 |
587 |
0.28 |
| chr2_129227304_129227665 | 0.69 |
A730036I17Rik |
RIKEN cDNA A730036I17 gene |
538 |
0.53 |
| chr7_97749255_97749938 | 0.67 |
Aqp11 |
aquaporin 11 |
11307 |
0.16 |
| chr2_29872842_29873027 | 0.67 |
2600006K01Rik |
RIKEN cDNA 2600006K01 gene |
2617 |
0.14 |
| chr13_55426059_55426382 | 0.64 |
F12 |
coagulation factor XII (Hageman factor) |
559 |
0.57 |
| chr6_90466371_90466910 | 0.63 |
Klf15 |
Kruppel-like factor 15 |
240 |
0.86 |
| chr7_28982862_28983600 | 0.63 |
Map4k1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
370 |
0.72 |
| chr14_87140347_87141190 | 0.62 |
Diaph3 |
diaphanous related formin 3 |
375 |
0.89 |
| chr15_78509793_78510375 | 0.59 |
Il2rb |
interleukin 2 receptor, beta chain |
14813 |
0.09 |
| chr11_33039362_33039684 | 0.59 |
Rpsa-ps4 |
ribosomal protein S4, pseudogene 4 |
180 |
0.95 |
| chr16_32671532_32671962 | 0.58 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
1407 |
0.33 |
| chr13_54687874_54688432 | 0.58 |
Rnf44 |
ring finger protein 44 |
35 |
0.96 |
| chr6_124463804_124465512 | 0.58 |
Clstn3 |
calsyntenin 3 |
136 |
0.92 |
| chr7_19022230_19023942 | 0.57 |
Foxa3 |
forkhead box A3 |
452 |
0.57 |
| chr2_105673131_105674553 | 0.55 |
Pax6 |
paired box 6 |
121 |
0.95 |
| chr2_30929161_30930147 | 0.54 |
Ptges |
prostaglandin E synthase |
209 |
0.9 |
| chr13_91262379_91262778 | 0.54 |
Gm4130 |
predicted gene 4130 |
160 |
0.95 |
| chr4_154603748_154603899 | 0.53 |
Gm13134 |
predicted gene 13134 |
2508 |
0.22 |
| chr4_63373706_63374431 | 0.50 |
Akna |
AT-hook transcription factor |
7135 |
0.11 |
| chr7_28755618_28756520 | 0.50 |
Ccer2 |
coiled-coil glutamate-rich protein 2 |
105 |
0.92 |
| chr3_17793835_17795104 | 0.50 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
427 |
0.75 |
| chr1_161766766_161767491 | 0.49 |
Gm5049 |
predicted gene 5049 |
21072 |
0.12 |
| chr14_121137043_121138056 | 0.47 |
Farp1 |
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) |
35470 |
0.2 |
| chr9_77756703_77756872 | 0.46 |
Gclc |
glutamate-cysteine ligase, catalytic subunit |
2252 |
0.23 |
| chr8_105606313_105606835 | 0.46 |
Ripor1 |
RHO family interacting cell polarization regulator 1 |
1319 |
0.23 |
| chr4_148593276_148593471 | 0.45 |
Srm |
spermidine synthase |
521 |
0.62 |
| chr7_4149072_4149804 | 0.45 |
Leng9 |
leukocyte receptor cluster (LRC) member 9 |
936 |
0.34 |
| chr7_45654411_45654773 | 0.44 |
Mamstr |
MEF2 activating motif and SAP domain containing transcriptional regulator |
10208 |
0.06 |
| chr13_42051086_42051757 | 0.43 |
Hivep1 |
human immunodeficiency virus type I enhancer binding protein 1 |
600 |
0.73 |
| chr4_149854254_149854922 | 0.43 |
Gm47301 |
predicted gene, 47301 |
36824 |
0.09 |
| chr9_102924868_102925148 | 0.43 |
Ryk |
receptor-like tyrosine kinase |
26453 |
0.15 |
| chr11_64010815_64011208 | 0.41 |
Gm12289 |
predicted gene 12289 |
1604 |
0.42 |
| chr19_55942541_55942745 | 0.40 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
44334 |
0.16 |
| chr2_118599857_118600417 | 0.40 |
Bub1b |
BUB1B, mitotic checkpoint serine/threonine kinase |
19 |
0.97 |
| chr4_141149281_141149432 | 0.40 |
Fbxo42 |
F-box protein 42 |
1434 |
0.29 |
| chr6_127119580_127119731 | 0.39 |
Gm43126 |
predicted gene 43126 |
3311 |
0.14 |
| chr14_79822704_79823014 | 0.39 |
Gm6999 |
predicted gene 6999 |
13853 |
0.16 |
| chr5_139324497_139325782 | 0.39 |
Adap1 |
ArfGAP with dual PH domains 1 |
483 |
0.72 |
| chr17_55878350_55878724 | 0.38 |
Zfp119a |
zinc finger protein 119a |
393 |
0.77 |
| chr2_125122649_125123534 | 0.37 |
Myef2 |
myelin basic protein expression factor 2, repressor |
323 |
0.84 |
| chr13_109631789_109631940 | 0.36 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
916 |
0.75 |
| chr13_23761268_23761510 | 0.35 |
H4c1 |
H4 clustered histone 1 |
159 |
0.77 |
| chrX_56597033_56598019 | 0.35 |
Mmgt1 |
membrane magnesium transporter 1 |
543 |
0.71 |
| chr3_36568782_36568933 | 0.35 |
Ccna2 |
cyclin A2 |
2012 |
0.21 |
| chr12_109455257_109457986 | 0.35 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
2426 |
0.16 |
| chr11_116657233_116658580 | 0.35 |
Prcd |
photoreceptor disc component |
750 |
0.43 |
| chr10_43739925_43740824 | 0.34 |
Gm40634 |
predicted gene, 40634 |
2713 |
0.19 |
| chr8_109737737_109738073 | 0.34 |
Atxn1l |
ataxin 1-like |
166 |
0.94 |
| chr16_42444471_42445397 | 0.33 |
Gap43 |
growth associated protein 43 |
104283 |
0.07 |
| chr17_35978858_35980224 | 0.33 |
Prr3 |
proline-rich polypeptide 3 |
15 |
0.78 |
| chr11_17952930_17953846 | 0.33 |
Etaa1 |
Ewing tumor-associated antigen 1 |
487 |
0.52 |
| chr9_124423028_124423645 | 0.32 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
247 |
0.91 |
| chrX_107815441_107816283 | 0.32 |
2610002M06Rik |
RIKEN cDNA 2610002M06 gene |
472 |
0.57 |
| chr8_13063174_13063450 | 0.31 |
Proz |
protein Z, vitamin K-dependent plasma glycoprotein |
2347 |
0.15 |
| chr5_110103230_110103381 | 0.31 |
Gtpbp6 |
GTP binding protein 6 (putative) |
2030 |
0.15 |
| chr11_113787418_113787592 | 0.31 |
Sdk2 |
sidekick cell adhesion molecule 2 |
22918 |
0.15 |
| chr11_121454005_121454156 | 0.31 |
Tbcd |
tubulin-specific chaperone d |
2131 |
0.23 |
| chr5_103654994_103656209 | 0.30 |
1700016H13Rik |
RIKEN cDNA 1700016H13 gene |
122 |
0.94 |
| chr7_16296336_16297433 | 0.30 |
Ccdc9 |
coiled-coil domain containing 9 |
10089 |
0.11 |
| chr5_123057629_123058391 | 0.30 |
Gm6444 |
predicted gene 6444 |
8232 |
0.09 |
| chr11_116130078_116131204 | 0.30 |
Trim65 |
tripartite motif-containing 65 |
483 |
0.63 |
| chr2_180892979_180894605 | 0.29 |
Mir124a-3 |
microRNA 124a-3 |
248 |
0.53 |
| chr2_69722536_69723698 | 0.28 |
Ppig |
peptidyl-prolyl isomerase G (cyclophilin G) |
24 |
0.96 |
| chr16_42635132_42635376 | 0.28 |
Gm49739 |
predicted gene, 49739 |
28672 |
0.25 |
| chr11_75173246_75173912 | 0.28 |
Mir132 |
microRNA 132 |
103 |
0.6 |
| chr1_68163528_68163679 | 0.28 |
Gm37100 |
predicted gene, 37100 |
47056 |
0.17 |
| chr5_65955085_65956246 | 0.28 |
4930480C01Rik |
RIKEN cDNA 4930480C01 gene |
4360 |
0.13 |
| chr2_4918566_4919706 | 0.28 |
Phyh |
phytanoyl-CoA hydroxylase |
56 |
0.96 |
| chr10_80002493_80003765 | 0.27 |
Abca7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
5514 |
0.08 |
| chr9_119321482_119322666 | 0.27 |
Oxsr1 |
oxidative-stress responsive 1 |
248 |
0.87 |
| chrX_105390628_105392456 | 0.27 |
5330434G04Rik |
RIKEN cDNA 5330434G04 gene |
212 |
0.93 |
| chr9_107612115_107612807 | 0.27 |
Sema3b |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
3232 |
0.08 |
| chr16_21796698_21797017 | 0.26 |
1300002E11Rik |
RIKEN cDNA 1300002E11 gene |
2465 |
0.17 |
| chr7_142094181_142095425 | 0.26 |
Dusp8 |
dual specificity phosphatase 8 |
469 |
0.54 |
| chr2_103604353_103604504 | 0.26 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
38118 |
0.14 |
| chr5_77140191_77140680 | 0.25 |
Chaer1 |
cardiac hypertrophy associated epigenetic regulator 1 |
183 |
0.93 |
| chr9_69364238_69364543 | 0.25 |
Gm15511 |
predicted gene 15511 |
143 |
0.96 |
| chr17_26537930_26538547 | 0.25 |
Gm50275 |
predicted gene, 50275 |
11 |
0.96 |
| chr3_94483546_94484940 | 0.24 |
Celf3 |
CUGBP, Elav-like family member 3 |
84 |
0.93 |
| chr4_137356552_137357603 | 0.24 |
Cdc42 |
cell division cycle 42 |
622 |
0.64 |
| chr1_79439702_79440415 | 0.23 |
Scg2 |
secretogranin II |
16 |
0.98 |
| chr1_36071298_36072369 | 0.23 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
3433 |
0.18 |
| chr2_153161085_153162219 | 0.23 |
Tm9sf4 |
transmembrane 9 superfamily protein member 4 |
104 |
0.96 |
| chr11_70237225_70237851 | 0.23 |
0610010K14Rik |
RIKEN cDNA 0610010K14 gene |
49 |
0.9 |
| chr1_59237625_59237950 | 0.23 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
556 |
0.74 |
| chr12_68997254_68997526 | 0.23 |
Gm47515 |
predicted gene, 47515 |
2420 |
0.27 |
| chr1_75168195_75168929 | 0.23 |
Zfand2b |
zinc finger, AN1 type domain 2B |
84 |
0.91 |
| chr2_122377464_122377895 | 0.23 |
Gm24409 |
predicted gene, 24409 |
7961 |
0.13 |
| chr13_83737592_83739114 | 0.23 |
Gm33366 |
predicted gene, 33366 |
182 |
0.66 |
| chr1_184851591_184852782 | 0.23 |
Mtarc2 |
mitochondrial amidoxime reducing component 2 |
5735 |
0.17 |
| chr9_22129665_22130147 | 0.22 |
Acp5 |
acid phosphatase 5, tartrate resistant |
692 |
0.42 |
| chr16_4639235_4639386 | 0.22 |
Vasn |
vasorin |
631 |
0.4 |
| chr2_120609365_120609854 | 0.22 |
Haus2 |
HAUS augmin-like complex, subunit 2 |
14 |
0.56 |
| chrX_101290098_101290258 | 0.22 |
Med12 |
mediator complex subunit 12 |
2956 |
0.13 |
| chr1_171375955_171376106 | 0.22 |
Nectin4 |
nectin cell adhesion molecule 4 |
5675 |
0.07 |
| chr7_127208929_127209633 | 0.22 |
Mylpf |
myosin light chain, phosphorylatable, fast skeletal muscle |
382 |
0.58 |
| chr2_157006331_157006656 | 0.22 |
Dsn1 |
DSN1 homolog, MIS12 kinetochore complex component |
81 |
0.95 |
| chr7_19932934_19933834 | 0.21 |
Igsf23 |
immunoglobulin superfamily, member 23 |
11514 |
0.07 |
| chr17_33760609_33760910 | 0.21 |
Rab11b |
RAB11B, member RAS oncogene family |
229 |
0.76 |
| chrX_16618092_16618532 | 0.21 |
Maoa |
monoamine oxidase A |
1386 |
0.49 |
| chr8_64921935_64922086 | 0.21 |
Gm45405 |
predicted gene 45405 |
10033 |
0.12 |
| chr1_79437543_79437779 | 0.21 |
Scg2 |
secretogranin II |
2381 |
0.37 |
| chr4_107185843_107185994 | 0.20 |
Tceanc2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
6802 |
0.13 |
| chr7_13123449_13124197 | 0.20 |
Vmn1r86 |
vomeronasal 1 receptor 86 |
3906 |
0.1 |
| chr5_129213898_129214049 | 0.20 |
Rps16-ps2 |
ribosomal protein S16, pseudogene 2 |
85456 |
0.08 |
| chr19_40611236_40612509 | 0.20 |
Tctn3 |
tectonic family member 3 |
316 |
0.59 |
| chr16_78375773_78376721 | 0.20 |
Btg3 |
BTG anti-proliferation factor 3 |
563 |
0.74 |
| chr5_53994852_53995991 | 0.20 |
Stim2 |
stromal interaction molecule 2 |
3078 |
0.32 |
| chr17_56474107_56476457 | 0.20 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
655 |
0.65 |
| chr3_105894723_105895429 | 0.19 |
Adora3 |
adenosine A3 receptor |
9345 |
0.12 |
| chr14_60384879_60385447 | 0.19 |
Amer2 |
APC membrane recruitment 2 |
6877 |
0.22 |
| chr5_103774852_103775003 | 0.19 |
Aff1 |
AF4/FMR2 family, member 1 |
9432 |
0.21 |
| chr8_84760342_84761249 | 0.19 |
Nfix |
nuclear factor I/X |
12601 |
0.1 |
| chr6_83053316_83054403 | 0.19 |
Htra2 |
HtrA serine peptidase 2 |
22 |
0.87 |
| chr11_35833437_35834524 | 0.19 |
Rars |
arginyl-tRNA synthetase |
481 |
0.78 |
| chr18_37800729_37801183 | 0.19 |
Pcdhgc3 |
protocadherin gamma subfamily C, 3 |
5408 |
0.06 |
| chr7_13278800_13279459 | 0.18 |
Lig1 |
ligase I, DNA, ATP-dependent |
136 |
0.77 |
| chr14_105926792_105926987 | 0.18 |
Spry2 |
sprouty RTK signaling antagonist 2 |
30070 |
0.19 |
| chr16_73223504_73223655 | 0.17 |
4930500H12Rik |
RIKEN cDNA 4930500H12 gene |
124591 |
0.06 |
| chr10_4491321_4491472 | 0.17 |
Gm22739 |
predicted gene, 22739 |
7162 |
0.17 |
| chr6_128525106_128525425 | 0.17 |
Pzp |
PZP, alpha-2-macroglobulin like |
1438 |
0.23 |
| chrX_106188534_106190478 | 0.17 |
Pgk1 |
phosphoglycerate kinase 1 |
2122 |
0.24 |
| chr12_110693597_110694119 | 0.17 |
Hsp90aa1 |
heat shock protein 90, alpha (cytosolic), class A member 1 |
434 |
0.74 |
| chr12_111943340_111944555 | 0.16 |
5033406O09Rik |
RIKEN cDNA 5033406O09 gene |
199 |
0.89 |
| chr16_75837887_75838038 | 0.16 |
Gm15554 |
predicted gene 15554 |
64905 |
0.09 |
| chr9_69397701_69398954 | 0.16 |
Ice2 |
interactor of little elongation complex ELL subunit 2 |
321 |
0.86 |
| chr10_81182846_81182997 | 0.16 |
Dapk3 |
death-associated protein kinase 3 |
84 |
0.87 |
| chr3_21935407_21936105 | 0.16 |
Gm43674 |
predicted gene 43674 |
62712 |
0.12 |
| chr8_88633608_88633759 | 0.16 |
Snx20 |
sorting nexin 20 |
2418 |
0.26 |
| chr9_88404072_88404710 | 0.16 |
Snx14 |
sorting nexin 14 |
5770 |
0.14 |
| chr1_132322108_132322259 | 0.16 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
5413 |
0.13 |
| chr4_134272765_134273261 | 0.16 |
Pdik1l |
PDLIM1 interacting kinase 1 like |
14212 |
0.08 |
| chr11_69979766_69980520 | 0.15 |
Elp5 |
elongator acetyltransferase complex subunit 5 |
68 |
0.82 |
| chr11_88180343_88181714 | 0.15 |
Cuedc1 |
CUE domain containing 1 |
1275 |
0.44 |
| chr17_29859997_29860447 | 0.15 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
9681 |
0.17 |
| chr1_61376694_61377204 | 0.14 |
9530026F06Rik |
RIKEN cDNA 9530026F06 gene |
1483 |
0.43 |
| chr2_59042712_59042879 | 0.14 |
Gm13557 |
predicted gene 13557 |
83088 |
0.08 |
| chr9_71976137_71976437 | 0.14 |
Gm37663 |
predicted gene, 37663 |
14984 |
0.1 |
| chr13_93939713_93939971 | 0.14 |
Gm24737 |
predicted gene, 24737 |
16468 |
0.17 |
| chr11_32281676_32281827 | 0.14 |
Hba-a1 |
hemoglobin alpha, adult chain 1 |
1760 |
0.2 |
| chr11_102632549_102633707 | 0.14 |
2810433D01Rik |
RIKEN cDNA 2810433D01 gene |
8712 |
0.1 |
| chr11_96629731_96630059 | 0.13 |
Skap1 |
src family associated phosphoprotein 1 |
78340 |
0.07 |
| chr11_55183909_55184222 | 0.13 |
Slc36a2 |
solute carrier family 36 (proton/amino acid symporter), member 2 |
1012 |
0.43 |
| chr11_54029637_54029788 | 0.13 |
Slc22a4 |
solute carrier family 22 (organic cation transporter), member 4 |
1622 |
0.32 |
| chr12_111946013_111946538 | 0.13 |
5033406O09Rik |
RIKEN cDNA 5033406O09 gene |
1791 |
0.21 |
| chr11_77872271_77873074 | 0.13 |
Pipox |
pipecolic acid oxidase |
10014 |
0.14 |
| chr8_12984359_12984568 | 0.13 |
Mcf2l |
mcf.2 transforming sequence-like |
167 |
0.92 |
| chr12_82469855_82470833 | 0.13 |
Gm5435 |
predicted gene 5435 |
26193 |
0.18 |
| chr18_86473556_86473849 | 0.13 |
Gm50384 |
predicted gene, 50384 |
227 |
0.95 |
| chr15_80078314_80078973 | 0.12 |
Snord83b |
small nucleolar RNA, C/D box 83B |
52 |
0.93 |
| chr10_81603027_81603178 | 0.12 |
Tle6 |
transducin-like enhancer of split 6 |
2029 |
0.14 |
| chr8_40510947_40511757 | 0.12 |
Cnot7 |
CCR4-NOT transcription complex, subunit 7 |
48 |
0.87 |
| chr2_90887830_90887981 | 0.12 |
C1qtnf4 |
C1q and tumor necrosis factor related protein 4 |
2045 |
0.18 |
| chr3_37422219_37422534 | 0.12 |
Spata5 |
spermatogenesis associated 5 |
1512 |
0.26 |
| chr8_126598039_126598520 | 0.12 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
4293 |
0.26 |
| chr10_81144225_81145434 | 0.12 |
Zbtb7a |
zinc finger and BTB domain containing 7a |
6876 |
0.07 |
| chr6_57702379_57703390 | 0.12 |
Lancl2 |
LanC (bacterial lantibiotic synthetase component C)-like 2 |
203 |
0.91 |
| chr5_137149473_137149929 | 0.11 |
Muc3 |
mucin 3, intestinal |
379 |
0.75 |
| chr8_94704045_94704876 | 0.11 |
Pllp |
plasma membrane proteolipid |
8182 |
0.12 |
| chr1_92473017_92473751 | 0.11 |
Ndufa10 |
NADH:ubiquinone oxidoreductase subunit A10 |
363 |
0.78 |
| chr1_165999821_165999972 | 0.11 |
Pou2f1 |
POU domain, class 2, transcription factor 1 |
2713 |
0.21 |
| chr2_32166741_32168109 | 0.11 |
Gm27805 |
predicted gene, 27805 |
5746 |
0.13 |
| chr5_107687686_107688880 | 0.11 |
5830411K02Rik |
RIKEN cDNA 5830411K02 gene |
112 |
0.7 |
| chr14_105071055_105071206 | 0.11 |
Gm5671 |
predicted gene 5671 |
27680 |
0.17 |
| chr2_122380394_122381168 | 0.11 |
Gm24409 |
predicted gene, 24409 |
4859 |
0.15 |
| chr3_116421092_116421666 | 0.10 |
Cdc14a |
CDC14 cell division cycle 14A |
1270 |
0.39 |
| chrX_8145862_8146216 | 0.10 |
Rbm3os |
RNA binding motif protein 3, opposite strand |
122 |
0.54 |
| chr11_90685580_90685876 | 0.10 |
Tom1l1 |
target of myb1-like 1 (chicken) |
1851 |
0.38 |
| chr13_19343314_19343465 | 0.10 |
Stard3nl |
STARD3 N-terminal like |
15736 |
0.2 |
| chr12_76676350_76677514 | 0.10 |
Sptb |
spectrin beta, erythrocytic |
33091 |
0.15 |
| chr6_124810954_124811934 | 0.10 |
Tpi1 |
triosephosphate isomerase 1 |
442 |
0.58 |
| chr2_105668422_105670370 | 0.10 |
Pax6 |
paired box 6 |
461 |
0.65 |
| chr17_33926039_33926463 | 0.10 |
Tapbp |
TAP binding protein |
133 |
0.83 |
| chr6_88041235_88041766 | 0.10 |
Gm44187 |
predicted gene, 44187 |
967 |
0.4 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.6 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.4 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.1 | 0.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.3 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.2 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.5 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.2 | GO:2000489 | regulation of hepatic stellate cell activation(GO:2000489) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0061643 | chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.0 | GO:0071029 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.0 | 0.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.0 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.3 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0080011 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.0 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.0 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |