Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
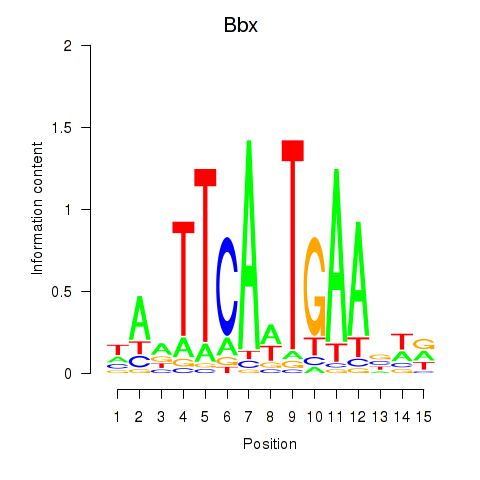
Results for Bbx
Z-value: 0.76
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSMUSG00000022641.9 | bobby sox HMG box containing |
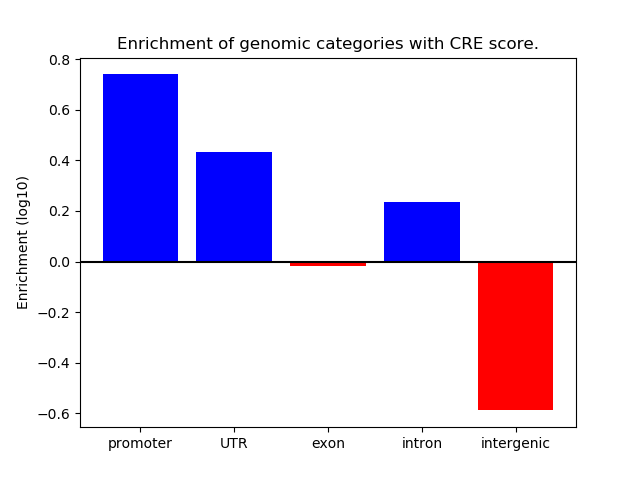
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_50363508_50364888 | Bbx | 33205 | 0.229547 | -0.35 | 5.8e-03 | Click! |
| chr16_50403156_50403361 | Bbx | 26654 | 0.243028 | -0.33 | 1.1e-02 | Click! |
| chr16_50367228_50367379 | Bbx | 36310 | 0.218872 | 0.31 | 1.5e-02 | Click! |
| chr16_50430464_50430776 | Bbx | 708 | 0.787579 | -0.27 | 3.4e-02 | Click! |
| chr16_50221557_50221752 | Bbx | 109339 | 0.068951 | -0.24 | 6.9e-02 | Click! |
Activity of the Bbx motif across conditions
Conditions sorted by the z-value of the Bbx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
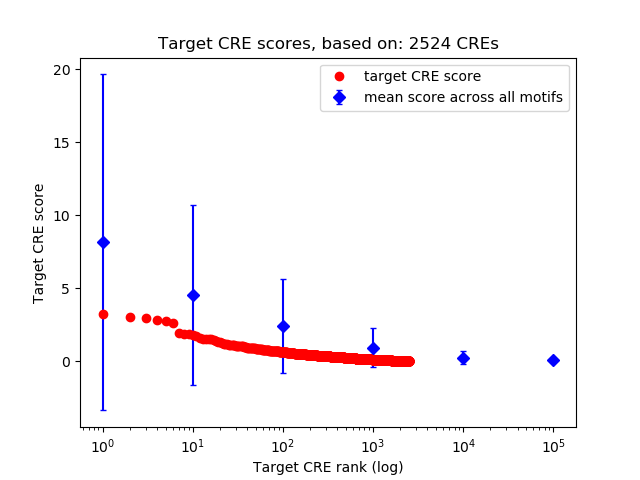
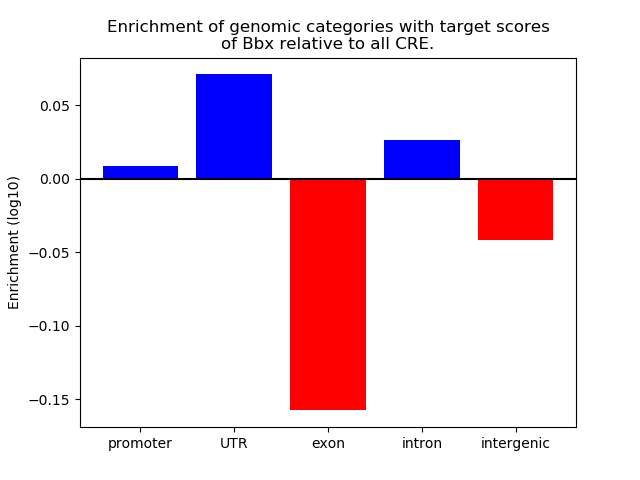
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_52699339_52699808 | 3.21 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
190 |
0.95 |
| chr11_41999400_42000640 | 3.04 |
Gabrg2 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
336 |
0.92 |
| chr8_54954519_54955779 | 2.96 |
Gpm6a |
glycoprotein m6a |
306 |
0.88 |
| chr12_29529828_29531185 | 2.84 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr13_83732205_83734272 | 2.75 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
672 |
0.58 |
| chr12_72234127_72234278 | 2.64 |
Rtn1 |
reticulon 1 |
1537 |
0.45 |
| chr9_58197310_58202560 | 1.92 |
Islr2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
637 |
0.54 |
| chr14_122479141_122479308 | 1.89 |
Zic2 |
zinc finger protein of the cerebellum 2 |
1124 |
0.34 |
| chr12_102555272_102555761 | 1.84 |
Chga |
chromogranin A |
530 |
0.74 |
| chr15_25026077_25026585 | 1.79 |
Gm2824 |
predicted gene 2824 |
1006 |
0.65 |
| chr5_97222612_97223731 | 1.76 |
Gm2861 |
predicted gene 2861 |
27585 |
0.16 |
| chr3_34654574_34655689 | 1.57 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
905 |
0.42 |
| chr2_53501543_53502209 | 1.55 |
Gm13503 |
predicted gene 13503 |
50050 |
0.17 |
| chr5_84413029_84413589 | 1.54 |
Epha5 |
Eph receptor A5 |
3497 |
0.31 |
| chr9_91404809_91406365 | 1.53 |
Gm29478 |
predicted gene 29478 |
1113 |
0.42 |
| chr6_15196934_15197697 | 1.50 |
Foxp2 |
forkhead box P2 |
351 |
0.94 |
| chr6_86031081_86032013 | 1.48 |
Add2 |
adducin 2 (beta) |
2800 |
0.16 |
| chr6_134886811_134888239 | 1.36 |
Gpr19 |
G protein-coupled receptor 19 |
243 |
0.87 |
| chr1_160350716_160351791 | 1.33 |
Rabgap1l |
RAB GTPase activating protein 1-like |
318 |
0.88 |
| chr3_34197711_34199105 | 1.30 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
1544 |
0.41 |
| chr2_124092543_124092695 | 1.28 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
2650 |
0.37 |
| chr2_34368351_34369364 | 1.19 |
Pbx3 |
pre B cell leukemia homeobox 3 |
1989 |
0.33 |
| chr12_46816152_46816702 | 1.18 |
Nova1 |
NOVA alternative splicing regulator 1 |
533 |
0.8 |
| chr17_78508180_78509392 | 1.18 |
Vit |
vitrin |
614 |
0.7 |
| chr14_124195406_124195650 | 1.14 |
Fgf14 |
fibroblast growth factor 14 |
2626 |
0.41 |
| chr19_38348426_38349048 | 1.12 |
Gm50150 |
predicted gene, 50150 |
6123 |
0.16 |
| chr6_55678280_55679200 | 1.12 |
Neurod6 |
neurogenic differentiation 6 |
2523 |
0.32 |
| chr4_43408488_43408639 | 1.12 |
Rusc2 |
RUN and SH3 domain containing 2 |
1234 |
0.34 |
| chr16_72510590_72511319 | 1.10 |
Robo1 |
roundabout guidance receptor 1 |
52746 |
0.18 |
| chr2_104406458_104406752 | 1.07 |
D430041D05Rik |
RIKEN cDNA D430041D05 gene |
3729 |
0.21 |
| chr1_99776348_99776679 | 1.06 |
Cntnap5b |
contactin associated protein-like 5B |
3748 |
0.27 |
| chr2_116053626_116054503 | 1.05 |
Meis2 |
Meis homeobox 2 |
4384 |
0.2 |
| chr19_47017205_47017356 | 1.03 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
2127 |
0.2 |
| chr9_104569742_104570707 | 1.02 |
Cpne4 |
copine IV |
437 |
0.89 |
| chr16_77418527_77418853 | 1.02 |
Gm38071 |
predicted gene, 38071 |
2066 |
0.19 |
| chr13_109442519_109443753 | 1.02 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
953 |
0.73 |
| chr16_77421091_77421970 | 1.01 |
9430053O09Rik |
RIKEN cDNA 9430053O09 gene |
290 |
0.84 |
| chr14_29718651_29719941 | 1.01 |
Cacna2d3 |
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
2499 |
0.26 |
| chr1_119833374_119833785 | 0.99 |
Ptpn4 |
protein tyrosine phosphatase, non-receptor type 4 |
3420 |
0.16 |
| chr2_181311130_181311281 | 0.91 |
Stmn3 |
stathmin-like 3 |
3295 |
0.12 |
| chr1_42693500_42693775 | 0.91 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
484 |
0.61 |
| chr1_176811621_176811894 | 0.91 |
Cep170 |
centrosomal protein 170 |
2310 |
0.18 |
| chr6_95574930_95575081 | 0.90 |
Suclg2 |
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
69004 |
0.13 |
| chr15_44751090_44751715 | 0.90 |
A930017M01Rik |
RIKEN cDNA A930017M01 gene |
878 |
0.47 |
| chr2_73776855_73777248 | 0.89 |
Chn1 |
chimerin 1 |
1705 |
0.39 |
| chr3_138745098_138745249 | 0.89 |
Tspan5 |
tetraspanin 5 |
2404 |
0.29 |
| chr18_79336296_79336940 | 0.89 |
Gm20593 |
predicted gene, 20593 |
11816 |
0.24 |
| chr7_136196592_136196867 | 0.89 |
Gm36737 |
predicted gene, 36737 |
23433 |
0.22 |
| chr10_20171692_20172198 | 0.88 |
Map7 |
microtubule-associated protein 7 |
62 |
0.97 |
| chr18_25750468_25751272 | 0.87 |
Celf4 |
CUGBP, Elav-like family member 4 |
1822 |
0.41 |
| chr8_102738030_102738197 | 0.86 |
Gm26301 |
predicted gene, 26301 |
5629 |
0.2 |
| chr9_40949123_40949736 | 0.86 |
Jhy |
junctional cadherin complex regulator |
12904 |
0.15 |
| chr3_107040280_107040795 | 0.85 |
AI504432 |
expressed sequence AI504432 |
1033 |
0.48 |
| chr4_102760289_102761654 | 0.84 |
Sgip1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
446 |
0.87 |
| chr3_45382505_45382699 | 0.83 |
Pcdh10 |
protocadherin 10 |
31 |
0.97 |
| chr1_168426195_168428871 | 0.82 |
Pbx1 |
pre B cell leukemia homeobox 1 |
3971 |
0.3 |
| chr9_41582824_41584205 | 0.82 |
Mir125b-1 |
microRNA 125b-1 |
1588 |
0.2 |
| chr4_23787162_23787456 | 0.81 |
Gm11890 |
predicted gene 11890 |
126965 |
0.06 |
| chr2_74713120_74713846 | 0.81 |
Hoxd3os1 |
homeobox D3, opposite strand 1 |
957 |
0.21 |
| chr6_55680954_55681113 | 0.80 |
Neurod6 |
neurogenic differentiation 6 |
230 |
0.94 |
| chr8_31912793_31913158 | 0.80 |
Nrg1 |
neuregulin 1 |
4675 |
0.26 |
| chr4_76445756_76446479 | 0.80 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
3863 |
0.26 |
| chr4_110284889_110285145 | 0.78 |
Elavl4 |
ELAV like RNA binding protein 4 |
1599 |
0.53 |
| chr14_45643431_45644460 | 0.77 |
Ddhd1 |
DDHD domain containing 1 |
11582 |
0.11 |
| chr13_73117045_73117937 | 0.77 |
Rpl31-ps2 |
ribosomal protein L31, pseudogene 2 |
115904 |
0.06 |
| chr1_41745210_41745391 | 0.77 |
Gm29260 |
predicted gene 29260 |
36508 |
0.23 |
| chr2_116070605_116071583 | 0.77 |
G630016G05Rik |
RIKEN cDNA G630016G05 gene |
3126 |
0.2 |
| chr16_6840364_6841436 | 0.77 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
31678 |
0.26 |
| chr5_124184090_124186568 | 0.76 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
211 |
0.9 |
| chr3_86916539_86917233 | 0.75 |
Dclk2 |
doublecortin-like kinase 2 |
3284 |
0.19 |
| chr2_116073034_116073185 | 0.73 |
2810405F15Rik |
RIKEN cDNA 2810405F15 gene |
2987 |
0.22 |
| chr2_14740186_14740953 | 0.72 |
Gm10848 |
predicted gene 10848 |
847 |
0.36 |
| chr9_56271539_56272112 | 0.72 |
Gm37842 |
predicted gene, 37842 |
8226 |
0.15 |
| chr1_25828771_25829000 | 0.71 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
626 |
0.54 |
| chr12_117157079_117158175 | 0.71 |
Gm10421 |
predicted gene 10421 |
5976 |
0.31 |
| chr4_13747506_13748460 | 0.71 |
Runx1t1 |
RUNX1 translocation partner 1 |
3314 |
0.37 |
| chr16_16558986_16560577 | 0.70 |
Fgd4 |
FYVE, RhoGEF and PH domain containing 4 |
209 |
0.94 |
| chr2_168763687_168764691 | 0.70 |
Sall4 |
spalt like transcription factor 4 |
1057 |
0.46 |
| chr5_134101199_134101480 | 0.69 |
Castor2 |
cytosolic arginine sensor for mTORC1 subunit 2 |
1334 |
0.34 |
| chr4_83500387_83501209 | 0.69 |
Gm11414 |
predicted gene 11414 |
37 |
0.96 |
| chr11_23495316_23495877 | 0.69 |
Ahsa2 |
AHA1, activator of heat shock protein ATPase 2 |
1942 |
0.18 |
| chr14_68119415_68119566 | 0.69 |
Nefm |
neurofilament, medium polypeptide |
5356 |
0.18 |
| chr13_84056577_84057434 | 0.68 |
Gm17750 |
predicted gene, 17750 |
7767 |
0.22 |
| chr11_32002849_32003000 | 0.68 |
Nsg2 |
neuron specific gene family member 2 |
2422 |
0.32 |
| chr3_80799469_80799764 | 0.68 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
2963 |
0.34 |
| chr12_72234504_72235243 | 0.68 |
Rtn1 |
reticulon 1 |
866 |
0.66 |
| chr9_72533965_72534704 | 0.67 |
Rfx7 |
regulatory factor X, 7 |
1599 |
0.21 |
| chr7_78881545_78881696 | 0.67 |
Mir7-2 |
microRNA 7-2 |
6657 |
0.12 |
| chr9_100686193_100686344 | 0.67 |
Gm38297 |
predicted gene, 38297 |
16945 |
0.16 |
| chr2_71775138_71775706 | 0.67 |
Itga6 |
integrin alpha 6 |
11507 |
0.13 |
| chr4_24429141_24429555 | 0.67 |
Gm27243 |
predicted gene 27243 |
1542 |
0.44 |
| chr2_97471929_97472576 | 0.67 |
Lrrc4c |
leucine rich repeat containing 4C |
4163 |
0.36 |
| chr3_86844695_86844863 | 0.66 |
Gm37025 |
predicted gene, 37025 |
19713 |
0.17 |
| chrX_57766985_57767599 | 0.66 |
Gm14631 |
predicted gene 14631 |
13424 |
0.18 |
| chr2_131351541_131352096 | 0.66 |
Rnf24 |
ring finger protein 24 |
1044 |
0.45 |
| chrX_99482036_99482258 | 0.66 |
Pja1 |
praja ring finger ubiquitin ligase 1 |
10874 |
0.2 |
| chr13_36728917_36729068 | 0.65 |
Nrn1 |
neuritin 1 |
1676 |
0.27 |
| chr9_91386509_91387874 | 0.65 |
Zic4 |
zinc finger protein of the cerebellum 4 |
4781 |
0.14 |
| chr11_96593909_96594964 | 0.64 |
Skap1 |
src family associated phosphoprotein 1 |
104575 |
0.05 |
| chr4_23636118_23636607 | 0.64 |
Gm25978 |
predicted gene, 25978 |
9617 |
0.24 |
| chr2_116052028_116053362 | 0.64 |
Meis2 |
Meis homeobox 2 |
3015 |
0.24 |
| chr16_77405996_77406237 | 0.64 |
Gm21816 |
predicted gene, 21816 |
5269 |
0.13 |
| chr1_79450112_79451070 | 0.63 |
Scg2 |
secretogranin II |
10471 |
0.24 |
| chr7_34809611_34810148 | 0.63 |
Chst8 |
carbohydrate sulfotransferase 8 |
2793 |
0.31 |
| chr10_100111299_100111726 | 0.63 |
Kitl |
kit ligand |
23456 |
0.13 |
| chr2_36094766_36095038 | 0.62 |
Lhx6 |
LIM homeobox protein 6 |
629 |
0.62 |
| chr2_6869935_6870654 | 0.62 |
Celf2 |
CUGBP, Elav-like family member 2 |
1678 |
0.39 |
| chr6_36807350_36807539 | 0.61 |
Ptn |
pleiotrophin |
2735 |
0.37 |
| chr14_118194572_118195025 | 0.61 |
4933431J24Rik |
RIKEN cDNA 4933431J24 gene |
14580 |
0.14 |
| chr5_146584081_146585559 | 0.61 |
Gpr12 |
G-protein coupled receptor 12 |
1 |
0.96 |
| chr16_91746458_91746610 | 0.60 |
Itsn1 |
intersectin 1 (SH3 domain protein 1A) |
9565 |
0.15 |
| chr18_37147091_37147753 | 0.59 |
Pcdhac2 |
protocadherin alpha subfamily C, 2 |
3919 |
0.14 |
| chr15_103026302_103028215 | 0.58 |
Hoxc4 |
homeobox C4 |
7137 |
0.09 |
| chr6_52105695_52105846 | 0.58 |
Halr1 |
Hoxa adjacent long noncoding RNA 1 |
2767 |
0.14 |
| chr2_118772778_118773205 | 0.57 |
Phgr1 |
proline/histidine/glycine-rich 1 |
222 |
0.88 |
| chrX_105389448_105389994 | 0.57 |
5330434G04Rik |
RIKEN cDNA 5330434G04 gene |
1807 |
0.32 |
| chr6_93911862_93913573 | 0.57 |
Magi1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
213 |
0.95 |
| chr10_86489595_86489929 | 0.57 |
Syn3 |
synapsin III |
2135 |
0.22 |
| chr11_77930982_77932135 | 0.57 |
Sez6 |
seizure related gene 6 |
519 |
0.68 |
| chr2_80870243_80870731 | 0.56 |
Gm13700 |
predicted gene 13700 |
8205 |
0.15 |
| chr14_48123838_48124177 | 0.56 |
Peli2 |
pellino 2 |
1978 |
0.25 |
| chr6_138424419_138425583 | 0.56 |
Lmo3 |
LIM domain only 3 |
386 |
0.83 |
| chr12_61527782_61528209 | 0.56 |
Lrfn5 |
leucine rich repeat and fibronectin type III domain containing 5 |
4047 |
0.23 |
| chr2_38342005_38342312 | 0.56 |
Lhx2 |
LIM homeobox protein 2 |
1066 |
0.44 |
| chr3_89327181_89327435 | 0.56 |
Efna3 |
ephrin A3 |
4343 |
0.08 |
| chr9_91227777_91228605 | 0.55 |
Gm29602 |
predicted gene 29602 |
12176 |
0.18 |
| chr16_7041535_7042315 | 0.55 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
27921 |
0.28 |
| chr7_130517404_130517739 | 0.55 |
Ate1 |
arginyltransferase 1 |
1890 |
0.25 |
| chrX_169837837_169837988 | 0.55 |
Mid1 |
midline 1 |
9753 |
0.25 |
| chr3_16940321_16940472 | 0.55 |
Gm26485 |
predicted gene, 26485 |
117084 |
0.07 |
| chr8_90956080_90956960 | 0.55 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
1085 |
0.49 |
| chr7_80187685_80187960 | 0.54 |
Sema4b |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
944 |
0.4 |
| chr4_19574638_19575636 | 0.54 |
Rmdn1 |
regulator of microtubule dynamics 1 |
25 |
0.98 |
| chr6_16727216_16727889 | 0.54 |
Gm36669 |
predicted gene, 36669 |
49972 |
0.17 |
| chr13_43215252_43215849 | 0.53 |
Tbc1d7 |
TBC1 domain family, member 7 |
44049 |
0.14 |
| chr1_50930540_50930933 | 0.53 |
Tmeff2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
3217 |
0.29 |
| chr4_126427336_126427559 | 0.53 |
Ago3 |
argonaute RISC catalytic subunit 3 |
2094 |
0.24 |
| chr18_42400574_42400820 | 0.53 |
Pou4f3 |
POU domain, class 4, transcription factor 3 |
5622 |
0.21 |
| chr14_60636814_60637375 | 0.52 |
Spata13 |
spermatogenesis associated 13 |
2339 |
0.35 |
| chr6_113991441_113991592 | 0.52 |
Gm15083 |
predicted gene 15083 |
13273 |
0.17 |
| chrX_166349290_166349906 | 0.52 |
Gpm6b |
glycoprotein m6b |
4756 |
0.26 |
| chr5_37241461_37244349 | 0.52 |
Crmp1 |
collapsin response mediator protein 1 |
171 |
0.95 |
| chr2_62088849_62089086 | 0.52 |
Slc4a10 |
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
42322 |
0.19 |
| chr14_64575544_64575771 | 0.52 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
11674 |
0.15 |
| chr2_79453025_79453285 | 0.52 |
Neurod1 |
neurogenic differentiation 1 |
3596 |
0.25 |
| chr5_83678355_83678676 | 0.51 |
Gm25806 |
predicted gene, 25806 |
11610 |
0.22 |
| chr9_80723838_80723989 | 0.51 |
Gm39380 |
predicted gene, 39380 |
13918 |
0.26 |
| chr1_42707054_42709031 | 0.51 |
Pantr2 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 2 |
10 |
0.97 |
| chr2_140668225_140669392 | 0.51 |
Flrt3 |
fibronectin leucine rich transmembrane protein 3 |
2592 |
0.43 |
| chr12_44439814_44440203 | 0.51 |
Nrcam |
neuronal cell adhesion molecule |
933 |
0.62 |
| chr8_47285815_47285966 | 0.51 |
Stox2 |
storkhead box 2 |
3472 |
0.3 |
| chr13_84905458_84906287 | 0.51 |
Gm4059 |
predicted gene 4059 |
68445 |
0.12 |
| chr12_119234321_119234603 | 0.51 |
Itgb8 |
integrin beta 8 |
4308 |
0.26 |
| chr11_96285767_96287533 | 0.50 |
Hoxb7 |
homeobox B7 |
27 |
0.93 |
| chr3_5225663_5226074 | 0.50 |
Zfhx4 |
zinc finger homeodomain 4 |
4363 |
0.22 |
| chr9_75681964_75682559 | 0.50 |
Scg3 |
secretogranin III |
1326 |
0.37 |
| chr10_87485570_87486803 | 0.50 |
Ascl1 |
achaete-scute family bHLH transcription factor 1 |
7474 |
0.2 |
| chr10_105201607_105201758 | 0.50 |
Tmtc2 |
transmembrane and tetratricopeptide repeat containing 2 |
1978 |
0.39 |
| chr11_30022183_30022992 | 0.50 |
Eml6 |
echinoderm microtubule associated protein like 6 |
3446 |
0.27 |
| chr11_98320492_98321552 | 0.49 |
Neurod2 |
neurogenic differentiation 2 |
8626 |
0.1 |
| chr13_78187267_78187418 | 0.49 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
3101 |
0.17 |
| chr12_107997791_107998940 | 0.49 |
Bcl11b |
B cell leukemia/lymphoma 11B |
5049 |
0.31 |
| chr3_124930856_124931558 | 0.49 |
Gm42825 |
predicted gene 42825 |
91698 |
0.09 |
| chr3_70482765_70483095 | 0.49 |
Gm6631 |
predicted gene 6631 |
69611 |
0.12 |
| chr5_63652583_63653162 | 0.49 |
Gm9954 |
predicted gene 9954 |
1978 |
0.33 |
| chr6_83113475_83113626 | 0.49 |
Mogs |
mannosyl-oligosaccharide glucosidase |
1946 |
0.1 |
| chr16_94372818_94373028 | 0.49 |
Pigp |
phosphatidylinositol glycan anchor biosynthesis, class P |
1081 |
0.39 |
| chrX_91163821_91163972 | 0.49 |
Gm14773 |
predicted gene 14773 |
8728 |
0.25 |
| chr11_103840627_103841312 | 0.49 |
Nsf |
N-ethylmaleimide sensitive fusion protein |
13538 |
0.18 |
| chr9_41587766_41588181 | 0.48 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
204 |
0.89 |
| chr6_107532409_107532560 | 0.48 |
Lrrn1 |
leucine rich repeat protein 1, neuronal |
2716 |
0.32 |
| chr17_78509585_78509946 | 0.48 |
Vit |
vitrin |
1593 |
0.34 |
| chr7_62416059_62417205 | 0.48 |
Mkrn3 |
makorin, ring finger protein, 3 |
3507 |
0.2 |
| chr1_166255711_166256817 | 0.47 |
Ildr2 |
immunoglobulin-like domain containing receptor 2 |
2071 |
0.3 |
| chr12_44438829_44439626 | 0.47 |
Nrcam |
neuronal cell adhesion molecule |
152 |
0.96 |
| chr3_146768414_146769205 | 0.47 |
Prkacb |
protein kinase, cAMP dependent, catalytic, beta |
1452 |
0.4 |
| chr19_44764612_44765334 | 0.47 |
Pax2 |
paired box 2 |
5058 |
0.15 |
| chr7_44312149_44312304 | 0.47 |
Shank1 |
SH3 and multiple ankyrin repeat domains 1 |
462 |
0.53 |
| chr3_4796453_4796708 | 0.47 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
978 |
0.63 |
| chr4_145625560_145625711 | 0.47 |
Gm13234 |
predicted gene 13234 |
6869 |
0.1 |
| chr9_60686843_60686994 | 0.46 |
Lrrc49 |
leucine rich repeat containing 49 |
616 |
0.74 |
| chr7_49842516_49843152 | 0.46 |
Gm44822 |
predicted gene 44822 |
28632 |
0.18 |
| chr17_6320795_6321910 | 0.46 |
AC183097.1 |
transmembrane protein 181 (TMEM181) pseudogene |
384 |
0.76 |
| chr8_87474515_87474666 | 0.46 |
Gm2694 |
predicted gene 2694 |
1196 |
0.32 |
| chr10_80382790_80383131 | 0.46 |
Gm22721 |
predicted gene, 22721 |
658 |
0.35 |
| chr6_136535739_136536498 | 0.46 |
Atf7ip |
activating transcription factor 7 interacting protein |
3191 |
0.19 |
| chr13_78196283_78196791 | 0.45 |
Nr2f1 |
nuclear receptor subfamily 2, group F, member 1 |
164 |
0.91 |
| chr8_61261964_61262115 | 0.45 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
26833 |
0.17 |
| chr13_15468099_15468909 | 0.45 |
Gli3 |
GLI-Kruppel family member GLI3 |
4524 |
0.2 |
| chr5_114586312_114586463 | 0.45 |
Fam222a |
family with sequence similarity 222, member A |
18370 |
0.14 |
| chr10_19397098_19397414 | 0.45 |
Olig3 |
oligodendrocyte transcription factor 3 |
40723 |
0.16 |
| chr4_142825883_142826722 | 0.45 |
Gm37624 |
predicted gene, 37624 |
35494 |
0.22 |
| chr4_6986587_6987701 | 0.45 |
Tox |
thymocyte selection-associated high mobility group box |
3339 |
0.35 |
| chr2_9886962_9888150 | 0.44 |
Gata3 |
GATA binding protein 3 |
228 |
0.87 |
| chr1_84693950_84694415 | 0.44 |
Mir5126 |
microRNA 5126 |
1657 |
0.28 |
| chr5_148391071_148391222 | 0.44 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
1669 |
0.44 |
| chr17_6489757_6490685 | 0.44 |
Tmem181b-ps |
transmembrane protein 181B, pseudogene |
23199 |
0.14 |
| chr17_9167368_9168623 | 0.44 |
6530411M01Rik |
RIKEN cDNA 6530411M01 gene |
159 |
0.96 |
| chr3_86542542_86543253 | 0.44 |
Lrba |
LPS-responsive beige-like anchor |
864 |
0.63 |
| chr8_84928375_84929627 | 0.44 |
Mast1 |
microtubule associated serine/threonine kinase 1 |
226 |
0.8 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.4 | 2.8 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.4 | GO:0098598 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.2 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.6 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 0.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.4 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.7 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.4 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.1 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.1 | 2.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.5 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.3 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0072069 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 1.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.2 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.1 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.1 | 0.8 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.2 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.1 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.0 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.2 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 1.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.1 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.5 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0070662 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0021604 | cranial nerve structural organization(GO:0021604) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.3 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.0 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:2000698 | positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.9 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 0.0 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 1.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 3.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 2.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 3.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 3.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.2 | GO:0016917 | G-protein coupled GABA receptor activity(GO:0004965) GABA receptor activity(GO:0016917) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.4 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 3.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0004119 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0052687 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |