Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
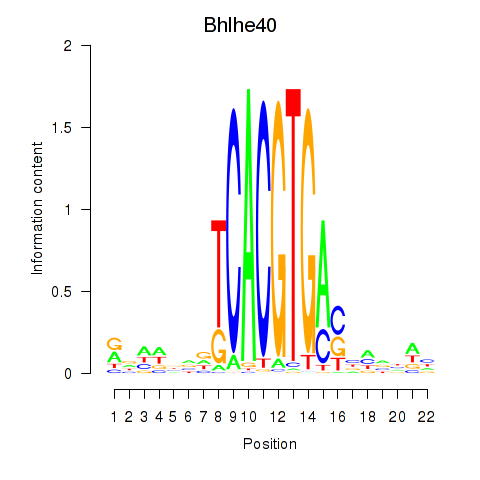
Results for Bhlhe40
Z-value: 0.48
Transcription factors associated with Bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bhlhe40
|
ENSMUSG00000030103.5 | basic helix-loop-helix family, member e40 |
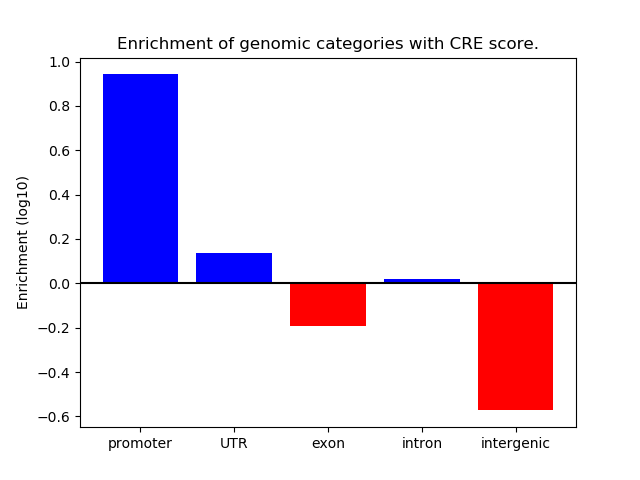
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_108708998_108710182 | Bhlhe40 | 46544 | 0.115962 | 0.49 | 7.6e-05 | Click! |
| chr6_108667829_108669053 | Bhlhe40 | 5395 | 0.178193 | 0.36 | 4.8e-03 | Click! |
| chr6_108659051_108660392 | Bhlhe40 | 908 | 0.412971 | 0.32 | 1.3e-02 | Click! |
| chr6_108700489_108700945 | Bhlhe40 | 37671 | 0.130362 | 0.30 | 2.1e-02 | Click! |
| chr6_108666033_108666205 | Bhlhe40 | 3073 | 0.213093 | -0.28 | 2.8e-02 | Click! |
Activity of the Bhlhe40 motif across conditions
Conditions sorted by the z-value of the Bhlhe40 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
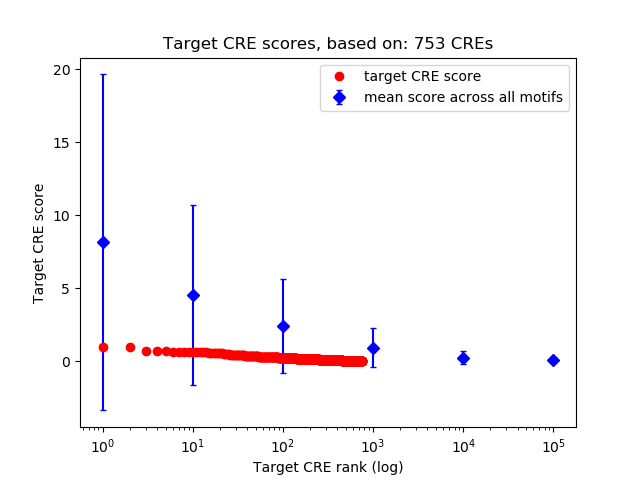
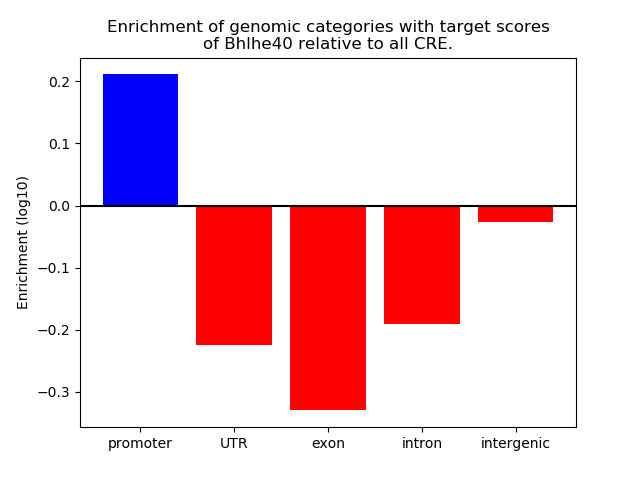
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_96328165_96328613 | 0.99 |
Hoxb3 |
homeobox B3 |
327 |
0.72 |
| chr6_145046367_145046518 | 0.98 |
Gm26666 |
predicted gene, 26666 |
1117 |
0.32 |
| chr9_40271177_40271411 | 0.71 |
Scn3b |
sodium channel, voltage-gated, type III, beta |
1607 |
0.28 |
| chr2_33633273_33633946 | 0.69 |
Lmx1b |
LIM homeobox transcription factor 1 beta |
1326 |
0.37 |
| chr10_117208235_117208395 | 0.68 |
Yeats4 |
YEATS domain containing 4 |
9043 |
0.11 |
| chr8_11729668_11729988 | 0.67 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
1651 |
0.22 |
| chr15_85676712_85677176 | 0.66 |
Lncppara |
long noncoding RNA near Ppara |
23328 |
0.12 |
| chr12_98215602_98215753 | 0.65 |
Galc |
galactosylceramidase |
851 |
0.61 |
| chr10_17796356_17797217 | 0.63 |
Txlnb |
taxilin beta |
530 |
0.7 |
| chr5_138194788_138195306 | 0.63 |
Mblac1 |
metallo-beta-lactamase domain containing 1 |
733 |
0.34 |
| chr2_74974235_74975165 | 0.63 |
n-R5s198 |
nuclear encoded rRNA 5S 198 |
39186 |
0.14 |
| chr13_48624936_48625515 | 0.62 |
Ptpdc1 |
protein tyrosine phosphatase domain containing 1 |
284 |
0.88 |
| chr1_78537079_78538149 | 0.61 |
Mogat1 |
monoacylglycerol O-acyltransferase 1 |
14 |
0.97 |
| chr14_19972733_19972884 | 0.60 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
3885 |
0.26 |
| chr9_77348040_77348200 | 0.60 |
Mlip |
muscular LMNA-interacting protein |
204 |
0.93 |
| chr13_66867979_66868968 | 0.60 |
Cbx3-ps4 |
chromobox 3, pseudogene 4 |
3949 |
0.09 |
| chrX_135210129_135210918 | 0.59 |
Tceal6 |
transcription elongation factor A (SII)-like 6 |
164 |
0.93 |
| chr13_65890336_65891205 | 0.55 |
Cbx3-ps1 |
chromobox 3, pseudogene 1 |
3989 |
0.13 |
| chr5_104046345_104046782 | 0.55 |
Nudt9 |
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
37 |
0.92 |
| chr12_72844062_72844595 | 0.54 |
Gm33785 |
predicted gene, 33785 |
26616 |
0.16 |
| chr7_46399823_46400899 | 0.53 |
Kcnc1 |
potassium voltage gated channel, Shaw-related subfamily, member 1 |
2713 |
0.23 |
| chr15_66283828_66283979 | 0.50 |
Kcnq3 |
potassium voltage-gated channel, subfamily Q, member 3 |
2148 |
0.3 |
| chr2_101677322_101677473 | 0.50 |
Traf6 |
TNF receptor-associated factor 6 |
1032 |
0.53 |
| chr14_123660983_123661518 | 0.49 |
Itgbl1 |
integrin, beta-like 1 |
174 |
0.96 |
| chr14_101886064_101886474 | 0.47 |
Lmo7 |
LIM domain only 7 |
2150 |
0.43 |
| chr7_29168805_29169490 | 0.46 |
Spred3 |
sprouty-related EVH1 domain containing 3 |
500 |
0.5 |
| chr13_83736071_83736534 | 0.45 |
Gm33366 |
predicted gene, 33366 |
2233 |
0.18 |
| chr4_34882920_34883280 | 0.45 |
Zfp292 |
zinc finger protein 292 |
140 |
0.94 |
| chr12_112653971_112654226 | 0.45 |
Akt1 |
thymoma viral proto-oncogene 1 |
2875 |
0.14 |
| chr8_114902581_114902732 | 0.44 |
Gm22556 |
predicted gene, 22556 |
150257 |
0.04 |
| chr5_110859375_110859531 | 0.43 |
Chek2 |
checkpoint kinase 2 |
6818 |
0.12 |
| chr13_44946654_44947258 | 0.43 |
Dtnbp1 |
dystrobrevin binding protein 1 |
188 |
0.96 |
| chr2_65848718_65849208 | 0.43 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
3108 |
0.27 |
| chr19_28967063_28967227 | 0.42 |
4430402I18Rik |
RIKEN cDNA 4430402I18 gene |
585 |
0.61 |
| chr1_178799792_178800387 | 0.41 |
Kif26b |
kinesin family member 26B |
1651 |
0.46 |
| chr2_65620767_65621991 | 0.41 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
568 |
0.82 |
| chr18_82765452_82765786 | 0.41 |
2210420H20Rik |
RIKEN cDNA 2210420H20 gene |
6578 |
0.12 |
| chr17_29503441_29503611 | 0.39 |
Pim1 |
proviral integration site 1 |
10119 |
0.1 |
| chrX_153831936_153833079 | 0.38 |
Spin2c |
spindlin family, member 2C |
214 |
0.94 |
| chr11_101552879_101553691 | 0.37 |
Nbr1 |
NBR1, autophagy cargo receptor |
384 |
0.68 |
| chr18_73863569_73864469 | 0.37 |
Mro |
maestro |
347 |
0.9 |
| chr15_25408150_25408416 | 0.37 |
Basp1 |
brain abundant, membrane attached signal protein 1 |
5415 |
0.17 |
| chr5_67444309_67444915 | 0.36 |
Gm20072 |
predicted gene, 20072 |
8867 |
0.13 |
| chr11_96306504_96308444 | 0.36 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
564 |
0.48 |
| chr13_65351676_65352548 | 0.35 |
Gm7762 |
predicted gene 7762 |
3515 |
0.09 |
| chr16_32645520_32645725 | 0.35 |
Tnk2 |
tyrosine kinase, non-receptor, 2 |
663 |
0.63 |
| chr4_76445756_76446479 | 0.35 |
Ptprd |
protein tyrosine phosphatase, receptor type, D |
3863 |
0.26 |
| chr1_191224508_191225332 | 0.35 |
D730003I15Rik |
RIKEN cDNA D730003I15 gene |
446 |
0.76 |
| chr11_74833809_74834108 | 0.34 |
Mnt |
max binding protein |
2368 |
0.2 |
| chr10_103367864_103369217 | 0.34 |
Slc6a15 |
solute carrier family 6 (neurotransmitter transporter), member 15 |
697 |
0.75 |
| chr4_140779841_140780227 | 0.33 |
Padi4 |
peptidyl arginine deiminase, type IV |
5798 |
0.14 |
| chr8_33833218_33834143 | 0.33 |
Rbpms |
RNA binding protein gene with multiple splicing |
9991 |
0.16 |
| chr5_136170674_136171277 | 0.33 |
Orai2 |
ORAI calcium release-activated calcium modulator 2 |
262 |
0.85 |
| chr1_87475065_87476008 | 0.33 |
Snorc |
secondary ossification center associated regulator of chondrocyte maturation |
2997 |
0.17 |
| chr6_143143889_143144415 | 0.32 |
Gm44306 |
predicted gene, 44306 |
9065 |
0.14 |
| chr1_40085123_40085879 | 0.32 |
Gm16894 |
predicted gene, 16894 |
350 |
0.57 |
| chr13_65420114_65420941 | 0.32 |
Cbx3-ps5 |
chromobox 3, pseudogene 5 |
3926 |
0.09 |
| chr6_37442520_37442875 | 0.32 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
551 |
0.84 |
| chr13_98012001_98012458 | 0.32 |
Arhgef28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
4188 |
0.31 |
| chr11_11461741_11462055 | 0.31 |
Spata48 |
spermatogenesis associated 48 |
196 |
0.78 |
| chr6_30572822_30572973 | 0.31 |
Gm13781 |
predicted gene 13781 |
3138 |
0.16 |
| chr5_125531759_125532074 | 0.31 |
Tmem132b |
transmembrane protein 132B |
142 |
0.95 |
| chr6_72097122_72097507 | 0.30 |
St3gal5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
278 |
0.85 |
| chr13_109568819_109569310 | 0.30 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
12341 |
0.32 |
| chrX_48256907_48257321 | 0.30 |
Utp14a |
UTP14A small subunit processome component |
196 |
0.93 |
| chrX_38772434_38772699 | 0.30 |
6030498E09Rik |
RIKEN cDNA 6030498E09 gene |
105 |
0.96 |
| chr3_126596101_126596252 | 0.30 |
Camk2d |
calcium/calmodulin-dependent protein kinase II, delta |
126 |
0.95 |
| chr2_26586309_26586605 | 0.30 |
Egfl7 |
EGF-like domain 7 |
173 |
0.48 |
| chr6_64688339_64689088 | 0.30 |
Grid2 |
glutamate receptor, ionotropic, delta 2 |
25525 |
0.22 |
| chr1_24588357_24588596 | 0.29 |
Col19a1 |
collagen, type XIX, alpha 1 |
1004 |
0.35 |
| chr1_143739622_143740266 | 0.29 |
Glrx2 |
glutaredoxin 2 (thioltransferase) |
298 |
0.87 |
| chr11_79254808_79254959 | 0.29 |
Wsb1 |
WD repeat and SOCS box-containing 1 |
212 |
0.93 |
| chr11_3863976_3864175 | 0.28 |
Osbp2 |
oxysterol binding protein 2 |
172 |
0.92 |
| chr13_41355674_41356040 | 0.28 |
Nedd9 |
neural precursor cell expressed, developmentally down-regulated gene 9 |
3390 |
0.18 |
| chr14_72603037_72603374 | 0.28 |
Fndc3a |
fibronectin type III domain containing 3A |
243 |
0.94 |
| chr8_91148783_91148985 | 0.28 |
Gm45665 |
predicted gene 45665 |
3323 |
0.14 |
| chr8_26258566_26258717 | 0.28 |
Gm31727 |
predicted gene, 31727 |
8336 |
0.13 |
| chr5_110102008_110102235 | 0.27 |
Plcxd1 |
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
917 |
0.36 |
| chr13_113031709_113032262 | 0.27 |
Cdc20b |
cell division cycle 20B |
3126 |
0.11 |
| chr11_96436700_96437020 | 0.27 |
Gm11529 |
predicted gene 11529 |
27687 |
0.11 |
| chr13_93612831_93613054 | 0.27 |
Gm15622 |
predicted gene 15622 |
12440 |
0.16 |
| chr19_14776871_14777022 | 0.27 |
Gm26026 |
predicted gene, 26026 |
58466 |
0.15 |
| chr4_118620317_118620501 | 0.27 |
Cfap57 |
cilia and flagella associated protein 57 |
4 |
0.85 |
| chr7_101897896_101898047 | 0.26 |
Anapc15 |
anaphase promoting complex C subunit 15 |
176 |
0.87 |
| chr16_25425048_25425199 | 0.26 |
Gm18896 |
predicted gene, 18896 |
18150 |
0.28 |
| chr8_85025253_85025568 | 0.26 |
Asna1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
129 |
0.86 |
| chr7_60960862_60961355 | 0.26 |
Gm44643 |
predicted gene 44643 |
277 |
0.94 |
| chr18_54910548_54911126 | 0.26 |
Zfp608 |
zinc finger protein 608 |
15275 |
0.19 |
| chr2_109955255_109955406 | 0.26 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
20212 |
0.18 |
| chr7_127376058_127376333 | 0.25 |
Zfp747 |
zinc finger protein 747 |
145 |
0.88 |
| chr15_40169641_40170367 | 0.25 |
Gm33301 |
predicted gene, 33301 |
7397 |
0.19 |
| chr1_84709872_84710637 | 0.25 |
Dner |
delta/notch-like EGF repeat containing |
14033 |
0.15 |
| chr17_25846144_25846295 | 0.25 |
Wdr90 |
WD repeat domain 90 |
275 |
0.68 |
| chr14_67608190_67608764 | 0.25 |
Gm47010 |
predicted gene, 47010 |
19685 |
0.17 |
| chr18_34572685_34572948 | 0.25 |
Nme5 |
NME/NM23 family member 5 |
6267 |
0.14 |
| chr12_108225542_108225693 | 0.25 |
Ccnk |
cyclin K |
26532 |
0.16 |
| chr9_63131800_63132681 | 0.24 |
Gm25064 |
predicted gene, 25064 |
1043 |
0.49 |
| chr17_87609597_87610246 | 0.24 |
Epcam |
epithelial cell adhesion molecule |
26058 |
0.15 |
| chr8_48224531_48225227 | 0.24 |
Gm32842 |
predicted gene, 32842 |
46106 |
0.16 |
| chr9_41950850_41951266 | 0.24 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
23529 |
0.17 |
| chr6_34175849_34176966 | 0.24 |
Slc35b4 |
solute carrier family 35, member B4 |
563 |
0.73 |
| chr10_81232757_81232928 | 0.24 |
Zfr2 |
zinc finger RNA binding protein 2 |
313 |
0.7 |
| chrX_136202895_136204183 | 0.24 |
Tceal5 |
transcription elongation factor A (SII)-like 5 |
98 |
0.94 |
| chr12_34221315_34221470 | 0.24 |
Gm18025 |
predicted gene, 18025 |
69743 |
0.13 |
| chr18_12168572_12168723 | 0.24 |
Rmc1 |
regulator of MON1-CCZ1 |
70 |
0.96 |
| chr2_94246278_94247531 | 0.23 |
Mir670hg |
MIR670 host gene (non-protein coding) |
3566 |
0.17 |
| chrX_134751410_134751672 | 0.23 |
Armcx6 |
armadillo repeat containing, X-linked 6 |
124 |
0.92 |
| chr7_47050631_47051464 | 0.23 |
Tmem86a |
transmembrane protein 86A |
446 |
0.55 |
| chr10_41519256_41519430 | 0.23 |
Cd164 |
CD164 antigen |
71 |
0.95 |
| chr16_90709814_90710620 | 0.23 |
Mis18a |
MIS18 kinetochore protein A |
16922 |
0.13 |
| chr13_85189150_85189414 | 0.23 |
Ccnh |
cyclin H |
126 |
0.97 |
| chr5_123603471_123603893 | 0.23 |
Clip1 |
CAP-GLY domain containing linker protein 1 |
37 |
0.96 |
| chr4_128654395_128654699 | 0.23 |
Phc2 |
polyhomeotic 2 |
155 |
0.95 |
| chr2_102448665_102449240 | 0.23 |
Fjx1 |
four jointed box 1 |
3547 |
0.29 |
| chr9_89622321_89623623 | 0.22 |
Minar1 |
membrane integral NOTCH2 associated receptor 1 |
153 |
0.95 |
| chr19_57473261_57473746 | 0.22 |
Trub1 |
TruB pseudouridine (psi) synthase family member 1 |
20546 |
0.13 |
| chrX_152326681_152327476 | 0.22 |
Kantr |
Kdm5c adjacent non-coding transcript |
385 |
0.81 |
| chr1_42918173_42918786 | 0.22 |
Mrps9 |
mitochondrial ribosomal protein S9 |
15422 |
0.18 |
| chr1_183296448_183297298 | 0.21 |
Brox |
BRO1 domain and CAAX motif containing |
119 |
0.58 |
| chr2_74670628_74670804 | 0.21 |
Hoxd13 |
homeobox D13 |
2406 |
0.09 |
| chr14_105565805_105566636 | 0.21 |
9330188P03Rik |
RIKEN cDNA 9330188P03 gene |
23256 |
0.16 |
| chr11_76409000_76409151 | 0.21 |
Timm22 |
translocase of inner mitochondrial membrane 22 |
1870 |
0.32 |
| chr11_70240091_70240373 | 0.21 |
Rnasek |
ribonuclease, RNase K |
390 |
0.4 |
| chr6_115837782_115838472 | 0.20 |
Efcab12 |
EF-hand calcium binding domain 12 |
285 |
0.81 |
| chr17_28517451_28518182 | 0.20 |
Fkbp5 |
FK506 binding protein 5 |
289 |
0.75 |
| chr14_57746100_57746612 | 0.20 |
Lats2 |
large tumor suppressor 2 |
222 |
0.52 |
| chr19_33761196_33761926 | 0.20 |
Lipo3 |
lipase, member O3 |
390 |
0.81 |
| chr8_93627469_93627620 | 0.20 |
Gm45708 |
predicted gene 45708 |
1080 |
0.55 |
| chr8_85024515_85025244 | 0.20 |
Asna1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
11 |
0.92 |
| chr14_54962885_54964094 | 0.20 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
3085 |
0.08 |
| chr19_17136519_17137673 | 0.20 |
Prune2 |
prune homolog 2 |
321 |
0.93 |
| chr16_16557820_16558051 | 0.20 |
Fgd4 |
FYVE, RhoGEF and PH domain containing 4 |
2055 |
0.32 |
| chr16_4211315_4211466 | 0.20 |
Crebbp |
CREB binding protein |
2023 |
0.29 |
| chr3_130730536_130730827 | 0.20 |
Gm42997 |
predicted gene 42997 |
80 |
0.75 |
| chr13_91223440_91223964 | 0.20 |
Atg10 |
autophagy related 10 |
257 |
0.91 |
| chr15_79400689_79401699 | 0.19 |
Tmem184b |
transmembrane protein 184b |
1725 |
0.23 |
| chr8_31089411_31091663 | 0.19 |
Dusp26 |
dual specificity phosphatase 26 (putative) |
733 |
0.65 |
| chr5_113082923_113083179 | 0.19 |
Crybb3 |
crystallin, beta B3 |
1467 |
0.26 |
| chr6_113341831_113342352 | 0.19 |
Camk1 |
calcium/calmodulin-dependent protein kinase I |
1174 |
0.25 |
| chr1_74376916_74377067 | 0.19 |
Slc11a1 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
334 |
0.78 |
| chr19_33589497_33590410 | 0.19 |
AC141640.1 |
renalase, FAD-dependent amine oxidase (Rnls) pseudogene |
54 |
0.82 |
| chr8_77717432_77717888 | 0.19 |
4933431K23Rik |
RIKEN cDNA 4933431K23 gene |
5924 |
0.19 |
| chr5_66336775_66338292 | 0.19 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
50 |
0.96 |
| chr6_145855324_145856436 | 0.19 |
Gm43909 |
predicted gene, 43909 |
7417 |
0.17 |
| chr2_142218264_142218415 | 0.19 |
Macrod2 |
mono-ADP ribosylhydrolase 2 |
41732 |
0.22 |
| chr18_76542618_76543294 | 0.19 |
Gm31933 |
predicted gene, 31933 |
89914 |
0.09 |
| chr12_3960025_3960885 | 0.19 |
Pomc |
pro-opiomelanocortin-alpha |
5485 |
0.16 |
| chr11_96382654_96382805 | 0.19 |
Gm11531 |
predicted gene 11531 |
9383 |
0.1 |
| chr2_59162876_59163950 | 0.19 |
Pkp4 |
plakophilin 4 |
1486 |
0.38 |
| chr13_112998291_112999283 | 0.18 |
Mcidas |
multiciliate differentiation and DNA synthesis associated cell cycle protein |
4942 |
0.11 |
| chr1_171033141_171033886 | 0.18 |
Gm26110 |
predicted gene, 26110 |
5548 |
0.1 |
| chr2_91068688_91068939 | 0.18 |
Slc39a13 |
solute carrier family 39 (metal ion transporter), member 13 |
58 |
0.95 |
| chr10_86685258_86685514 | 0.18 |
1810014B01Rik |
RIKEN cDNA 1810014B01 gene |
139 |
0.89 |
| chr17_66235333_66235831 | 0.18 |
Gm49932 |
predicted gene, 49932 |
637 |
0.64 |
| chr10_22731525_22732100 | 0.18 |
Tbpl1 |
TATA box binding protein-like 1 |
126 |
0.86 |
| chr5_113745200_113745351 | 0.18 |
Sart3 |
squamous cell carcinoma antigen recognized by T cells 3 |
320 |
0.82 |
| chr8_48740124_48740996 | 0.18 |
Tenm3 |
teneurin transmembrane protein 3 |
65870 |
0.13 |
| chr7_16311054_16311205 | 0.18 |
Bbc3 |
BCL2 binding component 3 |
674 |
0.57 |
| chr9_91365715_91366080 | 0.18 |
Zic1 |
zinc finger protein of the cerebellum 1 |
87 |
0.94 |
| chr2_84741934_84742491 | 0.18 |
Mir130a |
microRNA 130a |
1034 |
0.27 |
| chr10_33904707_33905726 | 0.18 |
Rsph4a |
radial spoke head 4 homolog A (Chlamydomonas) |
105 |
0.94 |
| chr9_55326354_55326761 | 0.18 |
Nrg4 |
neuregulin 4 |
192 |
0.69 |
| chr16_37916134_37917214 | 0.18 |
Gpr156 |
G protein-coupled receptor 156 |
178 |
0.94 |
| chr5_134932138_134932360 | 0.18 |
Mettl27 |
methyltransferase like 27 |
119 |
0.86 |
| chr6_100206731_100207328 | 0.17 |
Rybp |
RING1 and YY1 binding protein |
26102 |
0.16 |
| chr11_110379698_110380704 | 0.17 |
Map2k6 |
mitogen-activated protein kinase kinase 6 |
18921 |
0.23 |
| chr5_115373428_115373579 | 0.17 |
4930401G09Rik |
RIKEN cDNA 4930401G09 gene |
13380 |
0.07 |
| chr4_57039496_57040039 | 0.17 |
Epb41l4b |
erythrocyte membrane protein band 4.1 like 4b |
12091 |
0.21 |
| chr13_51570228_51570409 | 0.17 |
Shc3 |
src homology 2 domain-containing transforming protein C3 |
831 |
0.67 |
| chr6_92743458_92743787 | 0.17 |
Prickle2 |
prickle planar cell polarity protein 2 |
37467 |
0.18 |
| chr6_127717657_127718130 | 0.17 |
Gm42739 |
predicted gene 42739 |
45855 |
0.09 |
| chr10_85829040_85829963 | 0.17 |
Pwp1 |
PWP1 homolog, endonuclein |
7 |
0.75 |
| chr11_70970532_70970985 | 0.17 |
Rpain |
RPA interacting protein |
229 |
0.78 |
| chr4_80863119_80863395 | 0.17 |
Tyrp1 |
tyrosinase-related protein 1 |
16669 |
0.25 |
| chr6_122802291_122802442 | 0.17 |
Slc2a3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
726 |
0.49 |
| chr6_49214757_49215017 | 0.17 |
Igf2bp3 |
insulin-like growth factor 2 mRNA binding protein 3 |
70 |
0.97 |
| chr17_70561249_70561849 | 0.17 |
Dlgap1 |
DLG associated protein 1 |
3 |
0.99 |
| chr8_11730035_11730943 | 0.16 |
Arhgef7 |
Rho guanine nucleotide exchange factor (GEF7) |
2312 |
0.16 |
| chr14_61037991_61038513 | 0.16 |
Gm41168 |
predicted gene, 41168 |
292 |
0.5 |
| chr7_44973695_44975193 | 0.16 |
Cpt1c |
carnitine palmitoyltransferase 1c |
331 |
0.61 |
| chr11_120240839_120241468 | 0.16 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
4454 |
0.11 |
| chr5_124186580_124187887 | 0.16 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
51 |
0.96 |
| chr6_86438708_86439143 | 0.16 |
C87436 |
expressed sequence C87436 |
198 |
0.88 |
| chr13_67682587_67683510 | 0.16 |
Zfp738 |
zinc finger protein 738 |
419 |
0.69 |
| chr9_27791337_27792219 | 0.16 |
Opcml |
opioid binding protein/cell adhesion molecule-like |
476 |
0.89 |
| chr5_114090407_114091746 | 0.16 |
Svop |
SV2 related protein |
291 |
0.84 |
| chr15_68359591_68359932 | 0.16 |
Gm20732 |
predicted gene 20732 |
3415 |
0.16 |
| chr1_25825319_25826237 | 0.16 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
361 |
0.71 |
| chr11_53327066_53327217 | 0.16 |
Zcchc10 |
zinc finger, CCHC domain containing 10 |
259 |
0.88 |
| chr10_117417525_117417769 | 0.16 |
Gm40770 |
predicted gene, 40770 |
3743 |
0.17 |
| chr11_53692143_53692294 | 0.16 |
Rad50 |
RAD50 double strand break repair protein |
2782 |
0.16 |
| chr7_49119940_49120866 | 0.16 |
9130015G15Rik |
RIKEN cDNA 9130015G15 gene |
13873 |
0.19 |
| chr11_101086944_101087265 | 0.16 |
Mlx |
MAX-like protein X |
173 |
0.87 |
| chr2_44486358_44486509 | 0.16 |
Gm22867 |
predicted gene, 22867 |
57498 |
0.15 |
| chr12_29537800_29538885 | 0.16 |
Myt1l |
myelin transcription factor 1-like |
3120 |
0.28 |
| chr15_95652829_95653706 | 0.16 |
Dbx2 |
developing brain homeobox 2 |
2693 |
0.28 |
| chr4_134701681_134702469 | 0.16 |
Man1c1 |
mannosidase, alpha, class 1C, member 1 |
1692 |
0.39 |
| chr10_81722053_81722226 | 0.16 |
Zfp433 |
zinc finger protein 433 |
656 |
0.48 |
| chr17_25833142_25833995 | 0.16 |
Stub1 |
STIP1 homology and U-Box containing protein 1 |
164 |
0.8 |
| chr6_6577870_6578625 | 0.15 |
Sem1 |
SEM1, 26S proteasome complex subunit |
411 |
0.88 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.5 | GO:0009169 | ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.4 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.1 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.1 | 0.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.1 | 0.3 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.2 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.2 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:2000224 | regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.0 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.0 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.5 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.6 | GO:0052831 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |