Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
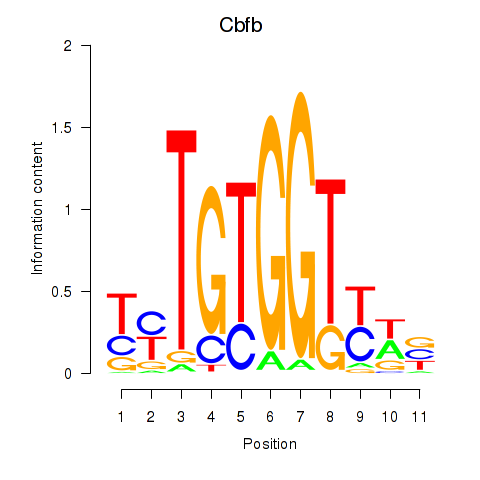
Results for Cbfb
Z-value: 0.34
Transcription factors associated with Cbfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cbfb
|
ENSMUSG00000031885.7 | core binding factor beta |
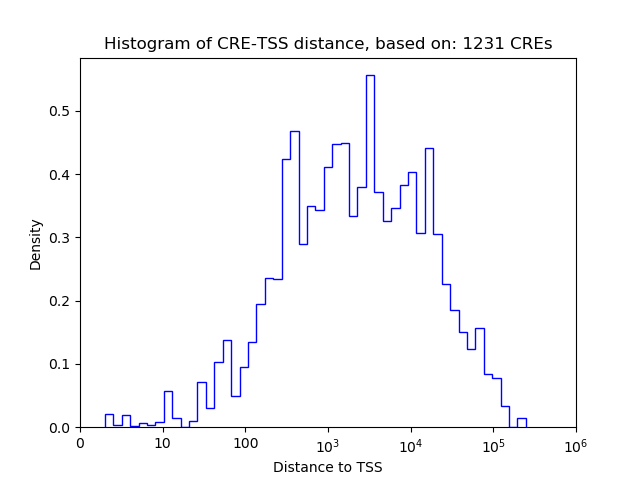
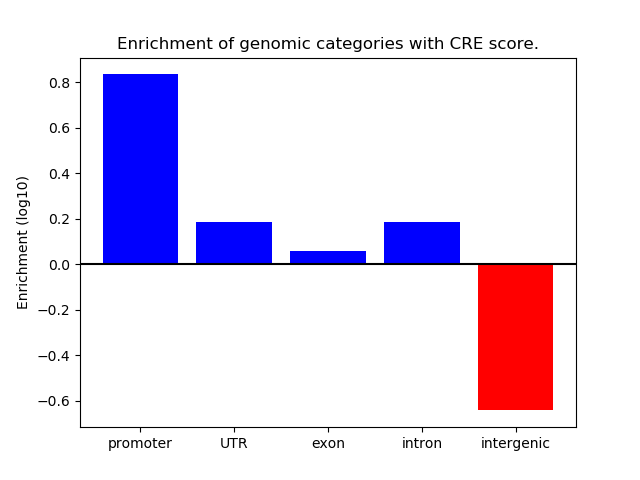
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_105169586_105169737 | Cbfb | 1013 | 0.332321 | -0.41 | 1.2e-03 | Click! |
| chr8_105170752_105171814 | Cbfb | 174 | 0.868766 | -0.22 | 8.5e-02 | Click! |
| chr8_105170135_105170411 | Cbfb | 401 | 0.699902 | 0.19 | 1.5e-01 | Click! |
Activity of the Cbfb motif across conditions
Conditions sorted by the z-value of the Cbfb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
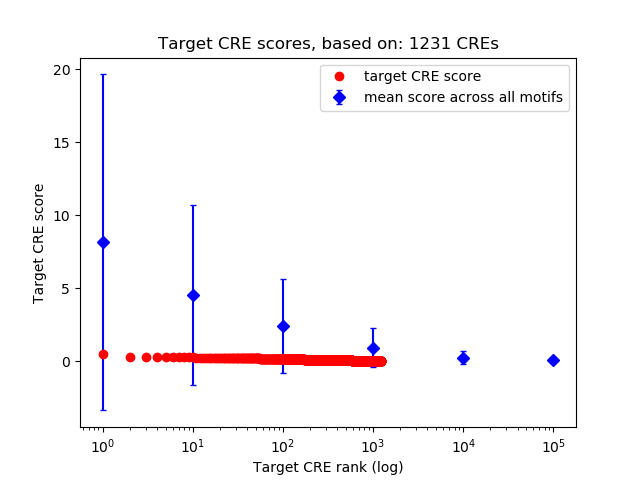
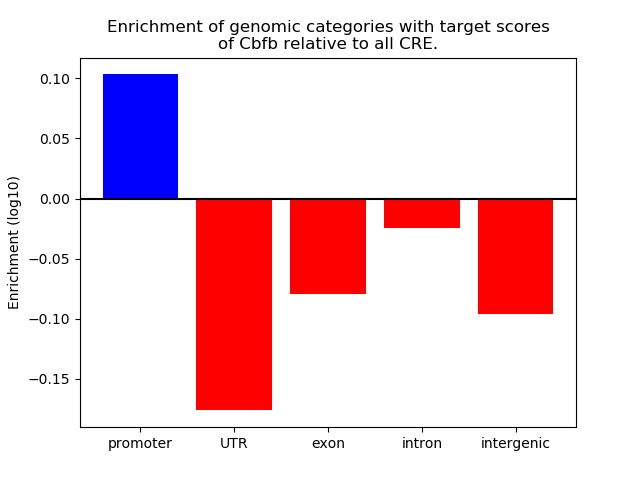
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_32167230_32167769 | 0.48 |
Ncoa4 |
nuclear receptor coactivator 4 |
1378 |
0.28 |
| chr11_83065104_83067047 | 0.32 |
Slfn2 |
schlafen 2 |
963 |
0.31 |
| chr4_49548394_49548727 | 0.31 |
Aldob |
aldolase B, fructose-bisphosphate |
986 |
0.46 |
| chr2_32081622_32082932 | 0.30 |
Fam78a |
family with sequence similarity 78, member A |
1506 |
0.26 |
| chr3_100485235_100486511 | 0.29 |
Tent5c |
terminal nucleotidyltransferase 5C |
3321 |
0.18 |
| chr6_90619935_90620303 | 0.28 |
Slc41a3 |
solute carrier family 41, member 3 |
972 |
0.47 |
| chr2_158145151_158146425 | 0.27 |
Tgm2 |
transglutaminase 2, C polypeptide |
578 |
0.71 |
| chr6_146220162_146220523 | 0.27 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
7201 |
0.26 |
| chr1_87620304_87621692 | 0.27 |
Inpp5d |
inositol polyphosphate-5-phosphatase D |
371 |
0.85 |
| chr2_27246009_27247328 | 0.26 |
Sardh |
sarcosine dehydrogenase |
180 |
0.94 |
| chr1_126603873_126604673 | 0.26 |
Nckap5 |
NCK-associated protein 5 |
111346 |
0.07 |
| chr11_96928897_96930218 | 0.25 |
Prr15l |
proline rich 15-like |
163 |
0.89 |
| chr2_180724979_180726144 | 0.25 |
Slc17a9 |
solute carrier family 17, member 9 |
161 |
0.92 |
| chr19_20389423_20390852 | 0.25 |
Anxa1 |
annexin A1 |
514 |
0.81 |
| chr7_75586786_75587711 | 0.25 |
Akap13 |
A kinase (PRKA) anchor protein 13 |
22791 |
0.17 |
| chr8_67948178_67948899 | 0.25 |
Psd3 |
pleckstrin and Sec7 domain containing 3 |
3341 |
0.28 |
| chr7_97748648_97749068 | 0.24 |
Aqp11 |
aquaporin 11 |
10569 |
0.16 |
| chr18_11058575_11059292 | 0.24 |
Gata6 |
GATA binding protein 6 |
114 |
0.97 |
| chrX_6579571_6580274 | 0.24 |
Shroom4 |
shroom family member 4 |
2636 |
0.42 |
| chr14_63270328_63271232 | 0.24 |
Gata4 |
GATA binding protein 4 |
344 |
0.87 |
| chr3_116860215_116860366 | 0.23 |
Frrs1 |
ferric-chelate reductase 1 |
723 |
0.56 |
| chr16_34922552_34923302 | 0.23 |
Mylk |
myosin, light polypeptide kinase |
7415 |
0.19 |
| chr15_41751298_41752572 | 0.23 |
Oxr1 |
oxidation resistance 1 |
234 |
0.95 |
| chr15_5107710_5108629 | 0.23 |
Card6 |
caspase recruitment domain family, member 6 |
315 |
0.81 |
| chr8_120292266_120293650 | 0.23 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
64502 |
0.09 |
| chr16_42907540_42907820 | 0.23 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
28 |
0.98 |
| chr2_127369985_127371247 | 0.23 |
Adra2b |
adrenergic receptor, alpha 2b |
7330 |
0.15 |
| chr7_105953249_105953965 | 0.23 |
Gm4070 |
predicted gene 4070 |
333 |
0.86 |
| chr7_106528440_106529206 | 0.22 |
Gm8995 |
predicted gene 8995 |
17152 |
0.19 |
| chr8_3353366_3353932 | 0.22 |
Arhgef18 |
rho/rac guanine nucleotide exchange factor (GEF) 18 |
189 |
0.95 |
| chr3_102165384_102165916 | 0.22 |
Vangl1 |
VANGL planar cell polarity 1 |
290 |
0.87 |
| chr19_5842574_5845856 | 0.22 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1044 |
0.25 |
| chr4_63153559_63154085 | 0.22 |
Ambp |
alpha 1 microglobulin/bikunin precursor |
351 |
0.86 |
| chr19_25052648_25052861 | 0.22 |
Dock8 |
dedicator of cytokinesis 8 |
141 |
0.97 |
| chr7_106214634_106215341 | 0.21 |
Gvin1 |
GTPase, very large interferon inducible 1 |
323 |
0.89 |
| chr9_63726302_63726832 | 0.21 |
Smad3 |
SMAD family member 3 |
14598 |
0.22 |
| chr6_115990945_115992684 | 0.21 |
Plxnd1 |
plexin D1 |
3191 |
0.2 |
| chr7_78914537_78914688 | 0.21 |
Isg20 |
interferon-stimulated protein |
283 |
0.86 |
| chr2_158306583_158307357 | 0.21 |
Lbp |
lipopolysaccharide binding protein |
358 |
0.8 |
| chr10_7472799_7473709 | 0.21 |
Ulbp1 |
UL16 binding protein 1 |
87 |
0.97 |
| chr5_38480596_38480837 | 0.21 |
Slc2a9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
410 |
0.84 |
| chr4_40851422_40852119 | 0.21 |
Gm25931 |
predicted gene, 25931 |
1368 |
0.21 |
| chr1_87636946_87637507 | 0.21 |
Inpp5d |
inositol polyphosphate-5-phosphatase D |
15552 |
0.16 |
| chr9_110709945_110711208 | 0.21 |
Ccdc12 |
coiled-coil domain containing 12 |
532 |
0.66 |
| chr6_38341600_38342409 | 0.20 |
Zc3hav1 |
zinc finger CCCH type, antiviral 1 |
12269 |
0.13 |
| chr11_79530296_79530704 | 0.20 |
Evi2 |
ecotropic viral integration site 2 |
59 |
0.52 |
| chr14_69278101_69278489 | 0.20 |
Gm20236 |
predicted gene, 20236 |
3845 |
0.11 |
| chr14_69496349_69496737 | 0.20 |
Gm37094 |
predicted gene, 37094 |
3847 |
0.12 |
| chr3_96830126_96830771 | 0.20 |
Pdzk1 |
PDZ domain containing 1 |
372 |
0.68 |
| chr11_29647613_29648153 | 0.20 |
Gm12092 |
predicted gene 12092 |
2374 |
0.23 |
| chr7_100854551_100855928 | 0.20 |
Relt |
RELT tumor necrosis factor receptor |
190 |
0.92 |
| chr6_125071043_125072167 | 0.20 |
Lpar5 |
lysophosphatidic acid receptor 5 |
328 |
0.72 |
| chr9_66157901_66159171 | 0.20 |
Dapk2 |
death-associated protein kinase 2 |
301 |
0.9 |
| chr5_35727461_35727612 | 0.20 |
Sh3tc1 |
SH3 domain and tetratricopeptide repeats 1 |
1476 |
0.36 |
| chr3_121547676_121548236 | 0.19 |
Slc44a3 |
solute carrier family 44, member 3 |
15552 |
0.16 |
| chr15_78844291_78845201 | 0.19 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
2122 |
0.16 |
| chr18_10532272_10533276 | 0.19 |
Gm24894 |
predicted gene, 24894 |
18632 |
0.16 |
| chr13_76581204_76581595 | 0.19 |
Mctp1 |
multiple C2 domains, transmembrane 1 |
1671 |
0.42 |
| chr6_48684916_48685953 | 0.19 |
Gimap4 |
GTPase, IMAP family member 4 |
852 |
0.34 |
| chr13_59821306_59822207 | 0.18 |
Tut7 |
terminal uridylyl transferase 7 |
832 |
0.41 |
| chrX_164421189_164421355 | 0.18 |
Piga |
phosphatidylinositol glycan anchor biosynthesis, class A |
1312 |
0.41 |
| chr19_21471462_21471819 | 0.18 |
Gda |
guanine deaminase |
948 |
0.67 |
| chr2_20736484_20738247 | 0.18 |
Etl4 |
enhancer trap locus 4 |
51 |
0.98 |
| chr8_13202008_13202379 | 0.18 |
2810030D12Rik |
RIKEN cDNA 2810030D12 gene |
1373 |
0.23 |
| chr19_53194082_53195694 | 0.18 |
Add3 |
adducin 3 (gamma) |
65 |
0.97 |
| chr5_137569837_137570642 | 0.18 |
Tfr2 |
transferrin receptor 2 |
370 |
0.67 |
| chr12_102355566_102356003 | 0.18 |
Rin3 |
Ras and Rab interactor 3 |
1004 |
0.59 |
| chr10_40133248_40133524 | 0.18 |
Slc16a10 |
solute carrier family 16 (monocarboxylic acid transporters), member 10 |
8868 |
0.14 |
| chr5_107724563_107727169 | 0.17 |
Gfi1 |
growth factor independent 1 transcription repressor |
61 |
0.88 |
| chr17_80564251_80564461 | 0.17 |
Cdkl4 |
cyclin-dependent kinase-like 4 |
522 |
0.78 |
| chrX_139611976_139612272 | 0.17 |
Rnf128 |
ring finger protein 128 |
757 |
0.7 |
| chr5_114378285_114378463 | 0.17 |
Kctd10 |
potassium channel tetramerisation domain containing 10 |
2109 |
0.21 |
| chr5_66114493_66114985 | 0.17 |
Rbm47 |
RNA binding motif protein 47 |
16548 |
0.11 |
| chr1_173741340_173741720 | 0.17 |
Ifi207 |
interferon activated gene 207 |
217 |
0.9 |
| chr5_123975536_123976239 | 0.17 |
Hip1r |
huntingtin interacting protein 1 related |
2259 |
0.17 |
| chr18_20552726_20553199 | 0.17 |
Tpi-rs10 |
triosephosphate isomerase related sequence 10 |
3598 |
0.2 |
| chr2_148441004_148443557 | 0.17 |
Cd93 |
CD93 antigen |
1283 |
0.41 |
| chr3_30766411_30766610 | 0.17 |
Samd7 |
sterile alpha motif domain containing 7 |
10288 |
0.14 |
| chr16_5007275_5008433 | 0.17 |
Smim22 |
small integral membrane protein 22 |
86 |
0.85 |
| chr16_34807078_34808442 | 0.16 |
Mylk |
myosin, light polypeptide kinase |
22839 |
0.23 |
| chr14_53795405_53795967 | 0.16 |
Trav12-4 |
T cell receptor alpha variable 12-4 |
17083 |
0.15 |
| chr15_50361344_50361976 | 0.16 |
Gm49198 |
predicted gene, 49198 |
74762 |
0.13 |
| chr13_56177742_56178675 | 0.16 |
Tifab |
TRAF-interacting protein with forkhead-associated domain, family member B |
593 |
0.68 |
| chr8_71558187_71559532 | 0.16 |
Tmem221 |
transmembrane protein 221 |
12 |
0.94 |
| chr16_23058440_23059079 | 0.16 |
Kng1 |
kininogen 1 |
366 |
0.69 |
| chr11_116615542_116615779 | 0.16 |
Rhbdf2 |
rhomboid 5 homolog 2 |
8540 |
0.1 |
| chr2_134825693_134826026 | 0.16 |
Gm14036 |
predicted gene 14036 |
21910 |
0.2 |
| chr10_111996341_111997155 | 0.16 |
Glipr1 |
GLI pathogenesis-related 1 (glioma) |
516 |
0.75 |
| chr19_32322600_32322981 | 0.16 |
Sgms1 |
sphingomyelin synthase 1 |
439 |
0.87 |
| chr19_34846228_34846905 | 0.16 |
Mir107 |
microRNA 107 |
25793 |
0.12 |
| chr13_56151379_56152016 | 0.16 |
Macroh2a1 |
macroH2A.1 histone |
15336 |
0.14 |
| chr4_57914999_57916744 | 0.16 |
D630039A03Rik |
RIKEN cDNA D630039A03 gene |
426 |
0.84 |
| chr12_116406570_116406928 | 0.16 |
Ncapg2 |
non-SMC condensin II complex, subunit G2 |
1315 |
0.32 |
| chr1_58975052_58975575 | 0.16 |
Stradb |
STE20-related kinase adaptor beta |
1791 |
0.24 |
| chr6_14752485_14752674 | 0.15 |
Ppp1r3a |
protein phosphatase 1, regulatory subunit 3A |
2695 |
0.42 |
| chr13_84651738_84651962 | 0.15 |
Gm26913 |
predicted gene, 26913 |
39091 |
0.22 |
| chr1_156615340_156616247 | 0.15 |
Abl2 |
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
130 |
0.96 |
| chr3_36606072_36606233 | 0.15 |
Bbs7 |
Bardet-Biedl syndrome 7 (human) |
7133 |
0.15 |
| chr4_53513691_53513842 | 0.15 |
Slc44a1 |
solute carrier family 44, member 1 |
21846 |
0.2 |
| chr10_78070047_78070198 | 0.15 |
Icosl |
icos ligand |
762 |
0.53 |
| chr13_19823886_19824322 | 0.15 |
Gpr141 |
G protein-coupled receptor 141 |
123 |
0.97 |
| chr6_115984719_115988278 | 0.15 |
Plxnd1 |
plexin D1 |
8507 |
0.15 |
| chr2_163507247_163507946 | 0.15 |
Hnf4a |
hepatic nuclear factor 4, alpha |
788 |
0.49 |
| chr18_56977658_56977824 | 0.15 |
C330018D20Rik |
RIKEN cDNA C330018D20 gene |
2373 |
0.33 |
| chr12_61093011_61093162 | 0.15 |
Gm48301 |
predicted gene, 48301 |
102040 |
0.08 |
| chr4_132794518_132794820 | 0.15 |
Themis2 |
thymocyte selection associated family member 2 |
1718 |
0.23 |
| chr16_38376199_38376921 | 0.15 |
Popdc2 |
popeye domain containing 2 |
4562 |
0.14 |
| chr2_127364227_127365175 | 0.15 |
Adra2b |
adrenergic receptor, alpha 2b |
1415 |
0.34 |
| chr9_42034159_42034310 | 0.15 |
Gm16214 |
predicted gene 16214 |
11619 |
0.22 |
| chr12_40381138_40381829 | 0.15 |
Zfp277 |
zinc finger protein 277 |
18599 |
0.22 |
| chr11_114072204_114072652 | 0.14 |
Sdk2 |
sidekick cell adhesion molecule 2 |
5382 |
0.24 |
| chr7_81122958_81123109 | 0.14 |
Slc28a1 |
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
8183 |
0.15 |
| chr2_163694593_163694992 | 0.14 |
Pkig |
protein kinase inhibitor, gamma |
754 |
0.55 |
| chr16_38350170_38350349 | 0.14 |
Cox17 |
cytochrome c oxidase assembly protein 17, copper chaperone |
967 |
0.41 |
| chr15_73751801_73752472 | 0.14 |
Ptp4a3 |
protein tyrosine phosphatase 4a3 |
3762 |
0.2 |
| chr6_99172298_99172685 | 0.14 |
Foxp1 |
forkhead box P1 |
9473 |
0.29 |
| chr18_75384437_75388058 | 0.14 |
Smad7 |
SMAD family member 7 |
11333 |
0.21 |
| chr2_109277608_109278317 | 0.14 |
Mettl15 |
methyltransferase like 15 |
318 |
0.9 |
| chr15_82792748_82793304 | 0.14 |
Cyp2d26 |
cytochrome P450, family 2, subfamily d, polypeptide 26 |
1196 |
0.3 |
| chr9_90237661_90237812 | 0.14 |
Gm16200 |
predicted gene 16200 |
16111 |
0.15 |
| chr17_83845850_83847094 | 0.14 |
Haao |
3-hydroxyanthranilate 3,4-dioxygenase |
277 |
0.9 |
| chr16_23028396_23028918 | 0.14 |
Kng2 |
kininogen 2 |
352 |
0.74 |
| chr16_20960637_20961170 | 0.14 |
Gm18769 |
predicted gene, 18769 |
239 |
0.91 |
| chr2_172393314_172394463 | 0.14 |
Cass4 |
Cas scaffolding protein family member 4 |
12 |
0.97 |
| chr4_47294508_47294837 | 0.14 |
Col15a1 |
collagen, type XV, alpha 1 |
6385 |
0.24 |
| chr15_95838442_95838593 | 0.14 |
Gm17546 |
predicted gene, 17546 |
8445 |
0.15 |
| chr8_104101710_104103631 | 0.14 |
Cdh5 |
cadherin 5 |
1045 |
0.43 |
| chr7_133700764_133701966 | 0.14 |
Uros |
uroporphyrinogen III synthase |
1173 |
0.35 |
| chr14_22020334_22020654 | 0.14 |
Lrmda |
leucine rich melanocyte differentiation associated |
646 |
0.58 |
| chr9_120116040_120116439 | 0.14 |
Slc25a38 |
solute carrier family 25, member 38 |
1250 |
0.23 |
| chr8_13202537_13203450 | 0.14 |
2810030D12Rik |
RIKEN cDNA 2810030D12 gene |
2173 |
0.16 |
| chr18_35739288_35740804 | 0.14 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
30 |
0.95 |
| chr1_173739285_173739489 | 0.14 |
Ifi207 |
interferon activated gene 207 |
2360 |
0.2 |
| chr4_118435800_118435951 | 0.14 |
Cdc20 |
cell division cycle 20 |
54 |
0.95 |
| chr12_85686314_85687645 | 0.14 |
Batf |
basic leucine zipper transcription factor, ATF-like |
310 |
0.85 |
| chr9_70385312_70385463 | 0.14 |
Mir5626 |
microRNA 5626 |
20366 |
0.17 |
| chr8_81012059_81012425 | 0.13 |
Gm9725 |
predicted gene 9725 |
1334 |
0.4 |
| chrX_133908388_133909247 | 0.13 |
Srpx2 |
sushi-repeat-containing protein, X-linked 2 |
369 |
0.84 |
| chr9_32698632_32699875 | 0.13 |
Ets1 |
E26 avian leukemia oncogene 1, 5' domain |
2869 |
0.25 |
| chr8_84197222_84198671 | 0.13 |
Gm26887 |
predicted gene, 26887 |
279 |
0.72 |
| chr13_12330029_12330316 | 0.13 |
Actn2 |
actinin alpha 2 |
10552 |
0.17 |
| chr1_130740681_130741391 | 0.13 |
Gm28857 |
predicted gene 28857 |
291 |
0.71 |
| chr5_67450455_67450649 | 0.13 |
Gm20072 |
predicted gene, 20072 |
2927 |
0.18 |
| chr9_44515217_44515909 | 0.13 |
Cxcr5 |
chemokine (C-X-C motif) receptor 5 |
10858 |
0.07 |
| chr7_100465236_100467118 | 0.13 |
C2cd3 |
C2 calcium-dependent domain containing 3 |
770 |
0.37 |
| chr5_105409514_105410008 | 0.13 |
Gm32051 |
predicted gene, 32051 |
394 |
0.82 |
| chr8_36961780_36962451 | 0.13 |
Dlc1 |
deleted in liver cancer 1 |
8972 |
0.19 |
| chr15_81043550_81043974 | 0.13 |
Mrtfa |
myocardin related transcription factor A |
1784 |
0.27 |
| chr14_79296843_79297470 | 0.13 |
Rgcc |
regulator of cell cycle |
4489 |
0.19 |
| chr2_84843620_84843993 | 0.13 |
Slc43a1 |
solute carrier family 43, member 1 |
3181 |
0.15 |
| chr13_42866202_42867352 | 0.13 |
Phactr1 |
phosphatase and actin regulator 1 |
57 |
0.98 |
| chrX_56446273_56446502 | 0.13 |
Gm2174 |
predicted gene 2174 |
1578 |
0.27 |
| chr7_19011655_19012657 | 0.13 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
8112 |
0.07 |
| chr3_105892115_105892622 | 0.13 |
Adora3 |
adenosine A3 receptor |
12053 |
0.11 |
| chr5_137631381_137632304 | 0.13 |
Lrch4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
1712 |
0.13 |
| chr14_75160166_75160317 | 0.13 |
Lcp1 |
lymphocyte cytosolic protein 1 |
14706 |
0.15 |
| chr2_35557994_35559313 | 0.13 |
Gm13446 |
predicted gene 13446 |
49 |
0.85 |
| chr5_134945425_134947032 | 0.13 |
Cldn4 |
claudin 4 |
706 |
0.44 |
| chr15_16315135_16315417 | 0.13 |
Gm6479 |
predicted gene 6479 |
107323 |
0.07 |
| chr8_89664040_89664191 | 0.13 |
Gm24212 |
predicted gene, 24212 |
3189 |
0.38 |
| chr8_79680146_79681210 | 0.13 |
Tpd52-ps |
tumor protein D52, pseudogene |
128 |
0.95 |
| chr2_65188482_65188857 | 0.13 |
Cobll1 |
Cobl-like 1 |
47326 |
0.14 |
| chr8_120831876_120832383 | 0.13 |
Gm26878 |
predicted gene, 26878 |
48077 |
0.12 |
| chr15_58594635_58595816 | 0.12 |
Fer1l6 |
fer-1-like 6 (C. elegans) |
43280 |
0.17 |
| chr8_104104538_104104713 | 0.12 |
Gm29682 |
predicted gene, 29682 |
1248 |
0.36 |
| chrX_74427105_74427296 | 0.12 |
Ikbkg |
inhibitor of kappaB kinase gamma |
702 |
0.45 |
| chr13_51737590_51738068 | 0.12 |
Sema4d |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
3066 |
0.26 |
| chr7_25774402_25774657 | 0.12 |
Axl |
AXL receptor tyrosine kinase |
149 |
0.91 |
| chr1_182928096_182928648 | 0.12 |
Gm37542 |
predicted gene, 37542 |
20662 |
0.18 |
| chr3_96223834_96224156 | 0.12 |
H2bc21 |
H2B clustered histone 21 |
2876 |
0.05 |
| chr1_78817332_78818790 | 0.12 |
Kcne4 |
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
1134 |
0.48 |
| chr9_113706351_113706642 | 0.12 |
Pdcd6ip |
programmed cell death 6 interacting protein |
1748 |
0.38 |
| chr4_141076285_141076997 | 0.12 |
Necap2 |
NECAP endocytosis associated 2 |
1584 |
0.24 |
| chrX_164373324_164374363 | 0.12 |
Vegfd |
vascular endothelial growth factor D |
465 |
0.82 |
| chr7_19266084_19266414 | 0.12 |
Vasp |
vasodilator-stimulated phosphoprotein |
297 |
0.76 |
| chr8_25542987_25545804 | 0.12 |
Gm16159 |
predicted gene 16159 |
9165 |
0.11 |
| chr13_100600285_100601357 | 0.12 |
Gm24261 |
predicted gene, 24261 |
3538 |
0.14 |
| chr10_120694138_120694963 | 0.12 |
4930471E19Rik |
RIKEN cDNA 4930471E19 gene |
4653 |
0.17 |
| chr7_145111605_145112276 | 0.12 |
Gm45181 |
predicted gene 45181 |
51056 |
0.14 |
| chr4_119894810_119895257 | 0.12 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
80538 |
0.1 |
| chr1_161734865_161735433 | 0.12 |
Gm15429 |
predicted pseudogene 15429 |
7660 |
0.15 |
| chr9_85323760_85324040 | 0.12 |
Tent5a |
terminal nucleotidyltransferase 5A |
3224 |
0.2 |
| chr3_68691756_68692227 | 0.12 |
Il12a |
interleukin 12a |
464 |
0.82 |
| chr3_19650548_19651144 | 0.12 |
Trim55 |
tripartite motif-containing 55 |
6338 |
0.16 |
| chr9_24485798_24486436 | 0.12 |
Dpy19l1 |
dpy-19-like 1 (C. elegans) |
1797 |
0.39 |
| chr4_106798811_106800382 | 0.12 |
Acot11 |
acyl-CoA thioesterase 11 |
194 |
0.93 |
| chr15_51918413_51918564 | 0.12 |
Gm48923 |
predicted gene, 48923 |
21401 |
0.15 |
| chr5_92525035_92525371 | 0.12 |
Scarb2 |
scavenger receptor class B, member 2 |
18370 |
0.14 |
| chr9_42461646_42462435 | 0.12 |
Tbcel |
tubulin folding cofactor E-like |
579 |
0.73 |
| chr18_32556111_32556702 | 0.12 |
Gypc |
glycophorin C |
3574 |
0.26 |
| chr2_35329508_35329682 | 0.12 |
Stom |
stomatin |
7381 |
0.14 |
| chr19_38042662_38044080 | 0.12 |
Myof |
myoferlin |
13 |
0.97 |
| chr12_7980336_7980771 | 0.12 |
Apob |
apolipoprotein B |
2774 |
0.29 |
| chrX_164141443_164141725 | 0.12 |
Ace2 |
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
1106 |
0.48 |
| chr17_35549775_35550249 | 0.11 |
Cdsn |
corneodesmosin |
2116 |
0.13 |
| chr18_13122372_13122612 | 0.11 |
Gm5240 |
predicted gene 5240 |
9069 |
0.22 |
| chr7_19696242_19697209 | 0.11 |
Apoe |
apolipoprotein E |
906 |
0.31 |
| chr10_45584015_45584201 | 0.11 |
Gm47504 |
predicted gene, 47504 |
2889 |
0.24 |
| chr4_141872806_141873468 | 0.11 |
Efhd2 |
EF hand domain containing 2 |
1783 |
0.2 |
| chr11_49076677_49076921 | 0.11 |
Ifi47 |
interferon gamma inducible protein 47 |
212 |
0.86 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.1 | 0.2 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0060264 | regulation of respiratory burst involved in inflammatory response(GO:0060264) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.0 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:1902996 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.0 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.0 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |