Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
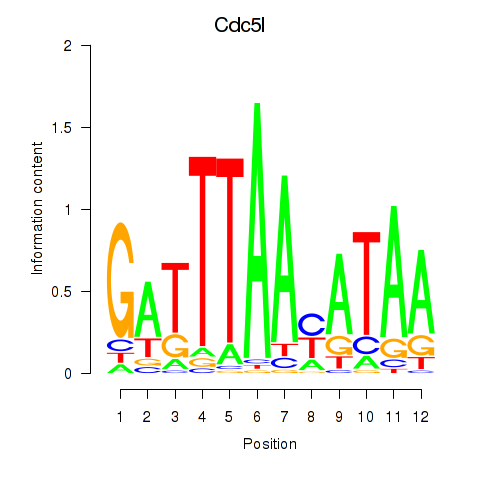
Results for Cdc5l
Z-value: 0.68
Transcription factors associated with Cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cdc5l
|
ENSMUSG00000023932.8 | cell division cycle 5-like (S. pombe) |
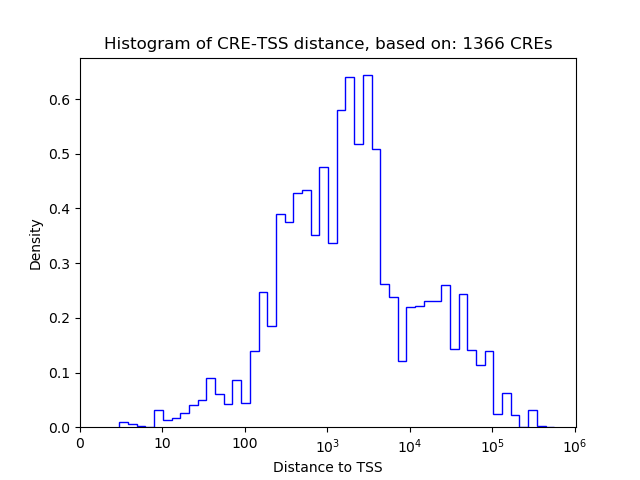
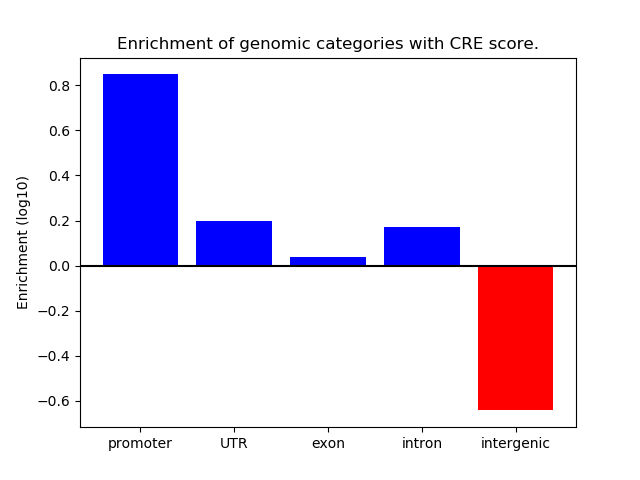
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_45432754_45433547 | Cdc5l | 391 | 0.502152 | 0.26 | 4.3e-02 | Click! |
| chr17_45361990_45362141 | Cdc5l | 53619 | 0.109625 | 0.20 | 1.3e-01 | Click! |
| chr17_45326704_45326930 | Cdc5l | 88867 | 0.065954 | 0.19 | 1.5e-01 | Click! |
| chr17_45318616_45318940 | Cdc5l | 96906 | 0.058838 | -0.18 | 1.8e-01 | Click! |
| chr17_45317989_45318514 | Cdc5l | 97433 | 0.058400 | -0.17 | 1.9e-01 | Click! |
Activity of the Cdc5l motif across conditions
Conditions sorted by the z-value of the Cdc5l motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
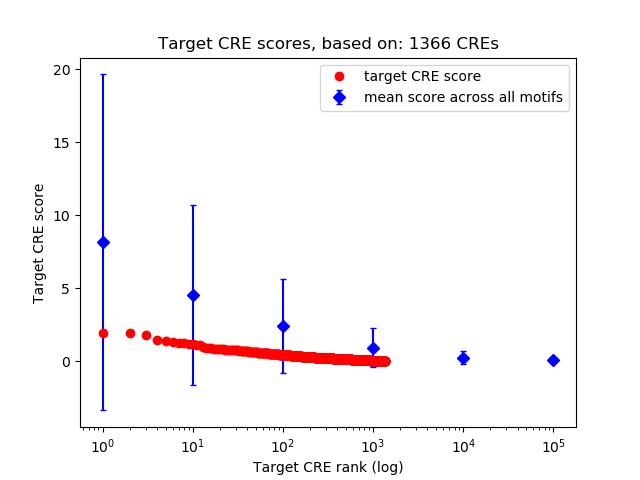
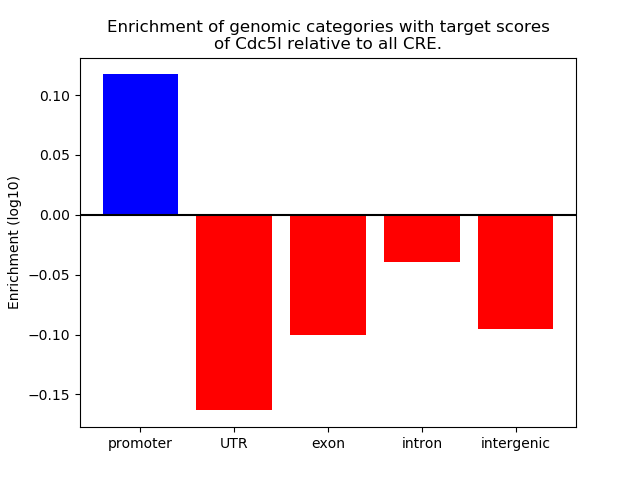
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_41585694_41587243 | 1.96 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
1301 |
0.29 |
| chr12_98572639_98572973 | 1.93 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
1906 |
0.26 |
| chr4_109343653_109343847 | 1.81 |
Eps15 |
epidermal growth factor receptor pathway substrate 15 |
497 |
0.8 |
| chr5_148265188_148266164 | 1.47 |
Mtus2 |
microtubule associated tumor suppressor candidate 2 |
331 |
0.91 |
| chr13_102957236_102957434 | 1.39 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
1011 |
0.69 |
| chr6_64799057_64799968 | 1.32 |
Atoh1 |
atonal bHLH transcription factor 1 |
70387 |
0.11 |
| chr12_70828137_70828628 | 1.27 |
Frmd6 |
FERM domain containing 6 |
2693 |
0.24 |
| chr12_29527051_29527810 | 1.23 |
Myt1l |
myelin transcription factor 1-like |
954 |
0.62 |
| chr10_69536775_69537233 | 1.22 |
Ank3 |
ankyrin 3, epithelial |
2782 |
0.32 |
| chr18_23036665_23037864 | 1.16 |
Nol4 |
nucleolar protein 4 |
1392 |
0.59 |
| chr8_65036696_65037878 | 1.15 |
Apela |
apelin receptor early endogenous ligand |
49 |
0.68 |
| chr6_36808050_36808542 | 1.11 |
Ptn |
pleiotrophin |
1883 |
0.46 |
| chr2_83814030_83814462 | 0.99 |
Fam171b |
family with sequence similarity 171, member B |
1610 |
0.34 |
| chr19_59460526_59461163 | 0.91 |
Emx2 |
empty spiracles homeobox 2 |
1325 |
0.36 |
| chr6_13835523_13837039 | 0.89 |
Gpr85 |
G protein-coupled receptor 85 |
960 |
0.59 |
| chr10_62863746_62864710 | 0.89 |
Tet1 |
tet methylcytosine dioxygenase 1 |
1076 |
0.37 |
| chr1_25228097_25229399 | 0.87 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
78 |
0.96 |
| chr3_27282892_27283771 | 0.85 |
Gm37191 |
predicted gene, 37191 |
5271 |
0.2 |
| chr10_109664804_109665193 | 0.83 |
3110043J17Rik |
RIKEN cDNA 3110043J17 gene |
16219 |
0.26 |
| chr13_116514424_116515376 | 0.82 |
Gm47913 |
predicted gene, 47913 |
148636 |
0.04 |
| chr7_30088185_30088336 | 0.81 |
Gm26920 |
predicted gene, 26920 |
2150 |
0.12 |
| chr18_37217058_37218378 | 0.81 |
Gm10544 |
predicted gene 10544 |
39196 |
0.08 |
| chrX_136115088_136116396 | 0.80 |
5730412P04Rik |
RIKEN cDNA 5730412P04 gene |
11139 |
0.12 |
| chr9_3403618_3403975 | 0.80 |
Cwf19l2 |
CWF19-like 2, cell cycle control (S. pombe) |
204 |
0.93 |
| chr18_13968574_13969239 | 0.78 |
Zfp521 |
zinc finger protein 521 |
2814 |
0.38 |
| chr6_136171003_136171483 | 0.77 |
Grin2b |
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
646 |
0.69 |
| chr14_52327318_52328620 | 0.76 |
Sall2 |
spalt like transcription factor 2 |
793 |
0.41 |
| chr3_57846307_57846818 | 0.76 |
Pfn2 |
profilin 2 |
583 |
0.72 |
| chr19_5797009_5797210 | 0.76 |
Malat1 |
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
157 |
0.88 |
| chr14_98164357_98165375 | 0.76 |
Dach1 |
dachshund family transcription factor 1 |
4677 |
0.28 |
| chr7_3390544_3391386 | 0.74 |
Cacng8 |
calcium channel, voltage-dependent, gamma subunit 8 |
282 |
0.73 |
| chr18_60925301_60926809 | 0.74 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
330 |
0.84 |
| chr2_63182035_63182719 | 0.74 |
Kcnh7 |
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
1793 |
0.52 |
| chr11_108926426_108927821 | 0.73 |
Axin2 |
axin 2 |
3942 |
0.24 |
| chr8_8656555_8657725 | 0.73 |
Efnb2 |
ephrin B2 |
4099 |
0.11 |
| chr2_105370584_105371218 | 0.72 |
Rcn1 |
reticulocalbin 1 |
24447 |
0.22 |
| chr18_70140801_70141850 | 0.72 |
Rab27b |
RAB27B, member RAS oncogene family |
249 |
0.95 |
| chr12_27340607_27340913 | 0.70 |
Sox11 |
SRY (sex determining region Y)-box 11 |
1814 |
0.48 |
| chr17_10309899_10310075 | 0.70 |
Qk |
quaking |
9374 |
0.24 |
| chr1_66323360_66324079 | 0.69 |
Map2 |
microtubule-associated protein 2 |
1617 |
0.37 |
| chr1_20893244_20893592 | 0.68 |
Paqr8 |
progestin and adipoQ receptor family member VIII |
2812 |
0.17 |
| chr3_84952327_84953098 | 0.66 |
Fbxw7 |
F-box and WD-40 domain protein 7 |
566 |
0.84 |
| chr1_168430152_168430657 | 0.66 |
Pbx1 |
pre B cell leukemia homeobox 1 |
1100 |
0.61 |
| chr13_45389099_45389552 | 0.65 |
Mylip |
myosin regulatory light chain interacting protein |
417 |
0.87 |
| chr14_58618020_58618171 | 0.64 |
Gm25614 |
predicted gene, 25614 |
32305 |
0.23 |
| chr3_154570067_154570517 | 0.64 |
Tyw3 |
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
24242 |
0.18 |
| chr16_63860769_63861945 | 0.64 |
Epha3 |
Eph receptor A3 |
2056 |
0.46 |
| chrX_69362591_69363214 | 0.64 |
Gm14705 |
predicted gene 14705 |
1633 |
0.4 |
| chr3_152264342_152264493 | 0.63 |
Nexn |
nexilin |
911 |
0.47 |
| chr13_78181011_78182180 | 0.62 |
Gm38604 |
predicted gene, 38604 |
1564 |
0.29 |
| chr6_33250271_33250473 | 0.61 |
Exoc4 |
exocyst complex component 4 |
1236 |
0.47 |
| chr19_19108280_19109261 | 0.61 |
Rorb |
RAR-related orphan receptor beta |
2426 |
0.42 |
| chr1_106174865_106175693 | 0.60 |
Phlpp1 |
PH domain and leucine rich repeat protein phosphatase 1 |
3527 |
0.22 |
| chr7_48847211_48847926 | 0.60 |
Csrp3 |
cysteine and glycine-rich protein 3 |
465 |
0.76 |
| chr18_82825279_82826286 | 0.58 |
4930445N18Rik |
RIKEN cDNA 4930445N18 gene |
30112 |
0.12 |
| chr12_26533196_26533974 | 0.58 |
Gm46344 |
predicted gene, 46344 |
46794 |
0.11 |
| chr10_88603199_88603736 | 0.58 |
Gm48752 |
predicted gene, 48752 |
995 |
0.39 |
| chr6_142506733_142508000 | 0.58 |
Ldhb |
lactate dehydrogenase B |
439 |
0.83 |
| chr16_44553746_44555096 | 0.57 |
Mir3081 |
microRNA 3081 |
3708 |
0.24 |
| chr9_61375691_61376224 | 0.57 |
Tle3 |
transducin-like enhancer of split 3 |
428 |
0.83 |
| chr11_77486623_77487566 | 0.56 |
Ankrd13b |
ankyrin repeat domain 13b |
2572 |
0.17 |
| chr12_3370750_3371679 | 0.56 |
Gm48511 |
predicted gene, 48511 |
2785 |
0.18 |
| chr4_126779227_126779591 | 0.56 |
Gm12937 |
predicted gene 12937 |
17006 |
0.12 |
| chr4_82496866_82497618 | 0.56 |
Nfib |
nuclear factor I/B |
2074 |
0.34 |
| chr3_74982117_74982268 | 0.55 |
Gm37464 |
predicted gene, 37464 |
42423 |
0.2 |
| chr14_76420544_76421824 | 0.54 |
Tsc22d1 |
TSC22 domain family, member 1 |
2356 |
0.39 |
| chr9_21616943_21617223 | 0.54 |
Smarca4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
819 |
0.44 |
| chr10_87491430_87491970 | 0.53 |
Ascl1 |
achaete-scute family bHLH transcription factor 1 |
1960 |
0.34 |
| chr3_9346361_9346885 | 0.53 |
C030034L19Rik |
RIKEN cDNA C030034L19 gene |
56441 |
0.13 |
| chr3_61366040_61366196 | 0.53 |
B430305J03Rik |
RIKEN cDNA B430305J03 gene |
167 |
0.94 |
| chr14_54617129_54617280 | 0.53 |
Mir686 |
microRNA 686 |
419 |
0.53 |
| chr9_60686646_60686797 | 0.52 |
Lrrc49 |
leucine rich repeat containing 49 |
813 |
0.64 |
| chr15_87545474_87545628 | 0.52 |
Tafa5 |
TAFA chemokine like family member 5 |
1252 |
0.64 |
| chr11_117873489_117873785 | 0.52 |
Tha1 |
threonine aldolase 1 |
156 |
0.9 |
| chr13_83728524_83729044 | 0.52 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
678 |
0.43 |
| chr8_90958597_90958859 | 0.52 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
3293 |
0.23 |
| chr12_86647308_86647551 | 0.51 |
Gm8504 |
predicted gene 8504 |
3144 |
0.21 |
| chr10_61625131_61626356 | 0.51 |
Gm28447 |
predicted gene 28447 |
14669 |
0.12 |
| chr12_59129648_59129986 | 0.51 |
Mia2 |
MIA SH3 domain ER export factor 2 |
7 |
0.97 |
| chr4_6987725_6988203 | 0.50 |
Tox |
thymocyte selection-associated high mobility group box |
2519 |
0.39 |
| chr5_127310557_127310877 | 0.50 |
9430087B13Rik |
RIKEN cDNA 9430087B13 gene |
1700 |
0.41 |
| chr14_108914298_108914499 | 0.50 |
Slitrk1 |
SLIT and NTRK-like family, member 1 |
240 |
0.96 |
| chr13_83987947_83988519 | 0.49 |
Gm4241 |
predicted gene 4241 |
242 |
0.93 |
| chr10_122044917_122045222 | 0.49 |
Srgap1 |
SLIT-ROBO Rho GTPase activating protein 1 |
2239 |
0.24 |
| chr6_86051805_86052622 | 0.48 |
Add2 |
adducin 2 (beta) |
35 |
0.96 |
| chr6_8352545_8353207 | 0.48 |
Gm16055 |
predicted gene 16055 |
11260 |
0.18 |
| chr12_36313868_36315206 | 0.48 |
Sostdc1 |
sclerostin domain containing 1 |
398 |
0.82 |
| chr18_43207273_43208441 | 0.48 |
Stk32a |
serine/threonine kinase 32A |
180 |
0.95 |
| chr19_28009042_28009226 | 0.47 |
Rfx3 |
regulatory factor X, 3 (influences HLA class II expression) |
1656 |
0.37 |
| chr5_49284738_49286021 | 0.47 |
Kcnip4 |
Kv channel interacting protein 4 |
280 |
0.92 |
| chr7_125393888_125394149 | 0.46 |
Gm30717 |
predicted gene, 30717 |
24096 |
0.14 |
| chr7_37689602_37690278 | 0.46 |
4930505M18Rik |
RIKEN cDNA 4930505M18 gene |
20141 |
0.2 |
| chr13_28949179_28949330 | 0.45 |
Sox4 |
SRY (sex determining region Y)-box 4 |
4459 |
0.24 |
| chr2_56454538_56455175 | 0.45 |
Mir195b |
microRNA 195b |
330955 |
0.01 |
| chr2_162658795_162659164 | 0.45 |
Ptprt |
protein tyrosine phosphatase, receptor type, T |
2115 |
0.26 |
| chr2_13175207_13176270 | 0.45 |
Gm37780 |
predicted gene, 37780 |
12115 |
0.2 |
| chr3_34562197_34562348 | 0.45 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
1880 |
0.28 |
| chr1_78192443_78193046 | 0.45 |
Pax3 |
paired box 3 |
4094 |
0.27 |
| chr13_67081589_67081868 | 0.44 |
Zfp708 |
zinc finger protein 708 |
183 |
0.87 |
| chr11_69561744_69562204 | 0.44 |
Efnb3 |
ephrin B3 |
1769 |
0.15 |
| chr1_126737352_126738681 | 0.44 |
Nckap5 |
NCK-associated protein 5 |
174 |
0.97 |
| chr7_27553482_27553949 | 0.44 |
2310022A10Rik |
RIKEN cDNA 2310022A10 gene |
429 |
0.47 |
| chr9_91369028_91370469 | 0.43 |
Zic4 |
zinc finger protein of the cerebellum 4 |
250 |
0.86 |
| chr7_16948034_16948663 | 0.43 |
Pnmal2 |
PNMA-like 2 |
3666 |
0.11 |
| chr4_124659206_124660160 | 0.43 |
Gm2164 |
predicted gene 2164 |
2514 |
0.16 |
| chr13_23544501_23545496 | 0.43 |
H3c7 |
H3 clustered histone 7 |
946 |
0.19 |
| chr10_42860045_42861117 | 0.43 |
Scml4 |
Scm polycomb group protein like 4 |
38 |
0.96 |
| chr14_19976049_19977076 | 0.43 |
Gng2 |
guanine nucleotide binding protein (G protein), gamma 2 |
131 |
0.97 |
| chr1_70725543_70726581 | 0.42 |
Vwc2l |
von Willebrand factor C domain-containing protein 2-like |
139 |
0.98 |
| chr5_117240468_117241490 | 0.42 |
Taok3 |
TAO kinase 3 |
519 |
0.72 |
| chr13_23561344_23562350 | 0.42 |
H3c6 |
H3 clustered histone 6 |
522 |
0.41 |
| chr11_34315006_34315399 | 0.41 |
Insyn2b |
inhibitory synaptic factor family member 2B |
380 |
0.85 |
| chr5_131532921_131534054 | 0.41 |
Auts2 |
autism susceptibility candidate 2 |
910 |
0.58 |
| chr7_73394818_73394989 | 0.41 |
Rgma |
repulsive guidance molecule family member A |
3627 |
0.16 |
| chr11_29189160_29189311 | 0.41 |
Ppp4r3b |
protein phosphatase 4 regulatory subunit 3B |
7952 |
0.18 |
| chr11_63914512_63915012 | 0.40 |
Hs3st3b1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
7528 |
0.23 |
| chr1_59684563_59684739 | 0.40 |
Nop58 |
NOP58 ribonucleoprotein |
320 |
0.78 |
| chr10_38967267_38967717 | 0.40 |
Lama4 |
laminin, alpha 4 |
1820 |
0.42 |
| chr16_41532916_41533606 | 0.40 |
Lsamp |
limbic system-associated membrane protein |
158 |
0.98 |
| chr7_90886512_90887836 | 0.40 |
Gm45159 |
predicted gene 45159 |
102 |
0.88 |
| chr7_46962989_46963387 | 0.40 |
Gm19248 |
predicted gene, 19248 |
4060 |
0.1 |
| chr8_121180685_121181292 | 0.39 |
5033426O07Rik |
RIKEN cDNA 5033426O07 gene |
20847 |
0.15 |
| chr1_23656325_23656702 | 0.39 |
Gm28822 |
predicted gene 28822 |
81926 |
0.09 |
| chr13_44841032_44841435 | 0.39 |
Jarid2 |
jumonji, AT rich interactive domain 2 |
450 |
0.83 |
| chr11_101628717_101628957 | 0.39 |
Rdm1 |
RAD52 motif 1 |
448 |
0.59 |
| chr2_19554051_19554670 | 0.39 |
4921504E06Rik |
RIKEN cDNA 4921504E06 gene |
446 |
0.82 |
| chr2_28792637_28792998 | 0.39 |
Gm13385 |
predicted gene 13385 |
752 |
0.58 |
| chr16_14705845_14706609 | 0.38 |
Snai2 |
snail family zinc finger 2 |
375 |
0.9 |
| chr10_77581317_77581531 | 0.38 |
Pttg1ip |
pituitary tumor-transforming 1 interacting protein |
296 |
0.82 |
| chr7_62407508_62407659 | 0.38 |
Mkrn3 |
makorin, ring finger protein, 3 |
12556 |
0.15 |
| chr6_53574522_53574673 | 0.38 |
Creb5 |
cAMP responsive element binding protein 5 |
1221 |
0.6 |
| chr8_69715849_69716479 | 0.38 |
Zfp869 |
zinc finger protein 869 |
290 |
0.87 |
| chr3_63962500_63962661 | 0.38 |
Gm26850 |
predicted gene, 26850 |
1053 |
0.38 |
| chr1_19212595_19213625 | 0.37 |
Tfap2b |
transcription factor AP-2 beta |
769 |
0.67 |
| chr9_101198368_101199080 | 0.37 |
Ppp2r3a |
protein phosphatase 2, regulatory subunit B'', alpha |
127 |
0.94 |
| chr14_114837724_114838180 | 0.37 |
4930524C18Rik |
RIKEN cDNA 4930524C18 gene |
5987 |
0.2 |
| chr9_22259919_22260226 | 0.37 |
Gm39307 |
predicted gene, 39307 |
30 |
0.67 |
| chr18_58206427_58207135 | 0.37 |
Fbn2 |
fibrillin 2 |
3145 |
0.33 |
| chr7_30165175_30165502 | 0.37 |
Zfp146 |
zinc finger protein 146 |
4353 |
0.08 |
| chr5_14025166_14026433 | 0.37 |
Sema3e |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
523 |
0.75 |
| chr3_56179928_56180616 | 0.37 |
Nbea |
neurobeachin |
3429 |
0.25 |
| chr1_178335228_178335429 | 0.37 |
Hnrnpu |
heterogeneous nuclear ribonucleoprotein U |
782 |
0.48 |
| chr13_83735558_83735982 | 0.36 |
Gm33366 |
predicted gene, 33366 |
2765 |
0.16 |
| chr2_84648361_84648948 | 0.36 |
Ctnnd1 |
catenin (cadherin associated protein), delta 1 |
117 |
0.92 |
| chr6_116107038_116107576 | 0.36 |
Gm20404 |
predicted gene 20404 |
343 |
0.63 |
| chr2_94184375_94184526 | 0.36 |
Gm23630 |
predicted gene, 23630 |
320 |
0.86 |
| chr10_111247804_111248910 | 0.36 |
Osbpl8 |
oxysterol binding protein-like 8 |
289 |
0.91 |
| chr8_48740124_48740996 | 0.36 |
Tenm3 |
teneurin transmembrane protein 3 |
65870 |
0.13 |
| chrX_75577044_75577513 | 0.36 |
Rab39b |
RAB39B, member RAS oncogene family |
953 |
0.4 |
| chr6_39874539_39874690 | 0.36 |
Tmem178b |
transmembrane protein 178B |
1543 |
0.32 |
| chr11_104158142_104158785 | 0.35 |
Crhr1 |
corticotropin releasing hormone receptor 1 |
25608 |
0.16 |
| chr5_126768981_126769769 | 0.35 |
Gm33347 |
predicted gene, 33347 |
42091 |
0.14 |
| chr9_67428325_67429235 | 0.35 |
Tln2 |
talin 2 |
23 |
0.98 |
| chr7_136007847_136008710 | 0.35 |
Gm9341 |
predicted gene 9341 |
55480 |
0.13 |
| chr14_84444241_84444671 | 0.35 |
Pcdh17 |
protocadherin 17 |
893 |
0.66 |
| chrX_135993531_135994683 | 0.35 |
Arxes2 |
adipocyte-related X-chromosome expressed sequence 2 |
283 |
0.88 |
| chr15_79030902_79031405 | 0.34 |
Gcat |
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
252 |
0.77 |
| chr4_22835212_22836357 | 0.33 |
Gm24078 |
predicted gene, 24078 |
88653 |
0.09 |
| chr15_98900276_98900427 | 0.33 |
Dhh |
desert hedgehog |
1811 |
0.14 |
| chr6_55680133_55680881 | 0.33 |
Neurod6 |
neurogenic differentiation 6 |
756 |
0.69 |
| chr2_94246278_94247531 | 0.33 |
Mir670hg |
MIR670 host gene (non-protein coding) |
3566 |
0.17 |
| chr3_45382505_45382699 | 0.33 |
Pcdh10 |
protocadherin 10 |
31 |
0.97 |
| chr6_112880016_112880276 | 0.33 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
50687 |
0.11 |
| chr15_85674019_85674170 | 0.32 |
Lncppara |
long noncoding RNA near Ppara |
20478 |
0.13 |
| chr2_104589693_104589847 | 0.32 |
Cstf3 |
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
753 |
0.54 |
| chr3_122462128_122462287 | 0.32 |
Gm25153 |
predicted gene, 25153 |
2520 |
0.2 |
| chr5_84414084_84414723 | 0.32 |
Epha5 |
Eph receptor A5 |
2403 |
0.37 |
| chr16_13357876_13359918 | 0.32 |
Mrtfb |
myocardin related transcription factor B |
476 |
0.83 |
| chr6_6857856_6858925 | 0.31 |
Gm44094 |
predicted gene, 44094 |
2139 |
0.2 |
| chr9_112118202_112119292 | 0.31 |
Mir128-2 |
microRNA 128-2 |
36 |
0.99 |
| chr4_9269280_9270516 | 0.31 |
Clvs1 |
clavesin 1 |
551 |
0.81 |
| chr3_144367127_144368131 | 0.31 |
Gm43447 |
predicted gene 43447 |
47415 |
0.13 |
| chr13_28811365_28811516 | 0.31 |
Gm17528 |
predicted gene, 17528 |
15683 |
0.19 |
| chr18_23038815_23040349 | 0.31 |
Nol4 |
nucleolar protein 4 |
496 |
0.88 |
| chr18_10787357_10787713 | 0.31 |
Mir133a-1hg |
Mir133a-1, Mir1b and Mir1a-2 host gene |
1652 |
0.2 |
| chr3_5225104_5225624 | 0.31 |
Zfhx4 |
zinc finger homeodomain 4 |
3859 |
0.22 |
| chr10_18548959_18549110 | 0.31 |
Hebp2 |
heme binding protein 2 |
2958 |
0.29 |
| chr12_29534253_29535510 | 0.30 |
Gm20208 |
predicted gene, 20208 |
10 |
0.8 |
| chr7_83077268_83077474 | 0.30 |
A530021J07Rik |
Riken cDNA A530021J07 gene |
7010 |
0.24 |
| chr11_116921808_116922404 | 0.30 |
Mgat5b |
mannoside acetylglucosaminyltransferase 5, isoenzyme B |
3243 |
0.21 |
| chr2_165599571_165599939 | 0.30 |
Gm28163 |
predicted gene 28163 |
2032 |
0.3 |
| chr11_50325927_50326447 | 0.30 |
Canx |
calnexin |
514 |
0.68 |
| chr18_31442020_31442171 | 0.30 |
Gm26658 |
predicted gene, 26658 |
1777 |
0.3 |
| chr16_77645925_77646470 | 0.30 |
Mir125b-2 |
microRNA 125b-2 |
76 |
0.58 |
| chr13_48264468_48265300 | 0.30 |
A330033J07Rik |
RIKEN cDNA A330033J07 gene |
2261 |
0.19 |
| chr12_118926946_118927231 | 0.30 |
Abcb5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
2508 |
0.38 |
| chr4_66025602_66025943 | 0.29 |
Gm11484 |
predicted gene 11484 |
328077 |
0.01 |
| chr6_113442321_113442500 | 0.29 |
Jagn1 |
jagunal homolog 1 |
159 |
0.85 |
| chr19_38482437_38482889 | 0.29 |
Plce1 |
phospholipase C, epsilon 1 |
1542 |
0.46 |
| chr19_28006304_28006455 | 0.29 |
Rfx3 |
regulatory factor X, 3 (influences HLA class II expression) |
4411 |
0.22 |
| chr18_20745187_20746004 | 0.29 |
B4galt6 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
809 |
0.66 |
| chr6_101198088_101199306 | 0.29 |
Gm26911 |
predicted gene, 26911 |
39 |
0.89 |
| chr1_182412012_182412163 | 0.28 |
Trp53bp2 |
transformation related protein 53 binding protein 2 |
2915 |
0.19 |
| chr8_92133986_92134310 | 0.28 |
Gm45332 |
predicted gene 45332 |
24118 |
0.17 |
| chr17_4907323_4907483 | 0.28 |
4930579D07Rik |
RIKEN cDNA 4930579D07 gene |
14416 |
0.19 |
| chr19_50676608_50676776 | 0.28 |
Gm26629 |
predicted gene, 26629 |
714 |
0.64 |
| chr13_83717521_83718816 | 0.28 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
3213 |
0.17 |
| chr14_64576826_64577138 | 0.28 |
Mir124a-1hg |
Mir124-1 host gene (non-protein coding) |
10349 |
0.16 |
| chr14_57744536_57744714 | 0.28 |
Lats2 |
large tumor suppressor 2 |
29 |
0.96 |
| chr17_24209746_24209897 | 0.28 |
Ntn3 |
netrin 3 |
290 |
0.71 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 0.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.6 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 0.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.5 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.4 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.5 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.1 | 0.4 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.5 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.3 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 | 0.5 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 0.4 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 0.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.2 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 1.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.4 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.3 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.2 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0071692 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.2 | GO:1900084 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.0 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:1900113 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.2 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.0 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.0 | 0.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.3 | GO:1902668 | negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.0 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0071107 | response to parathyroid hormone(GO:0071107) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.5 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0004487 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.0 | GO:0070815 | procollagen-lysine 5-dioxygenase activity(GO:0008475) peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |