Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
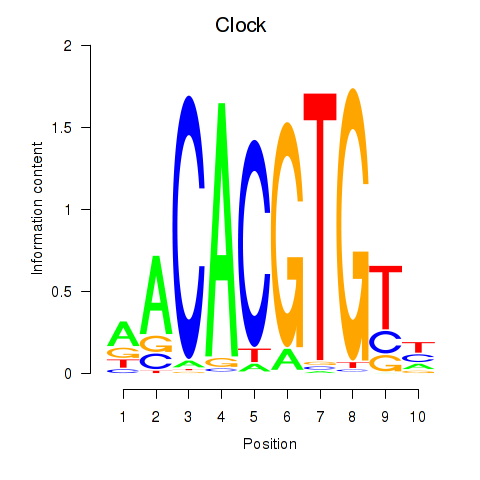
Results for Clock
Z-value: 0.65
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSMUSG00000029238.8 | circadian locomotor output cycles kaput |
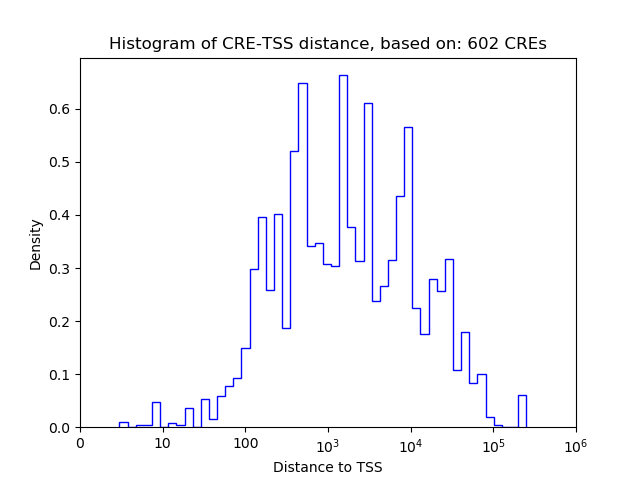
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_76303673_76304282 | Clock | 571 | 0.499670 | -0.12 | 3.6e-01 | Click! |
| chr5_76304304_76305311 | Clock | 15 | 0.534725 | 0.01 | 9.3e-01 | Click! |
Activity of the Clock motif across conditions
Conditions sorted by the z-value of the Clock motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
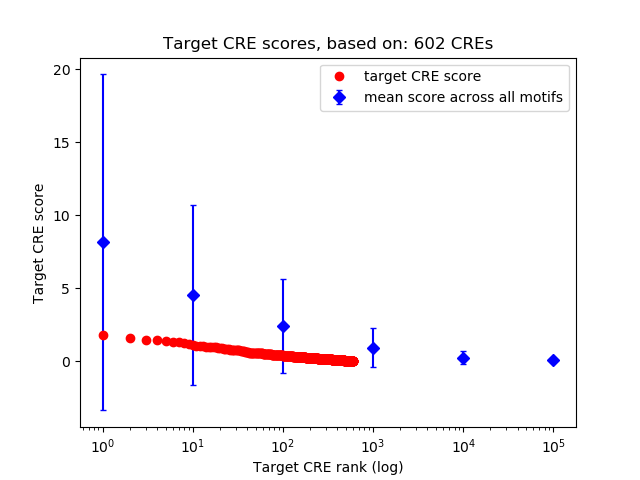
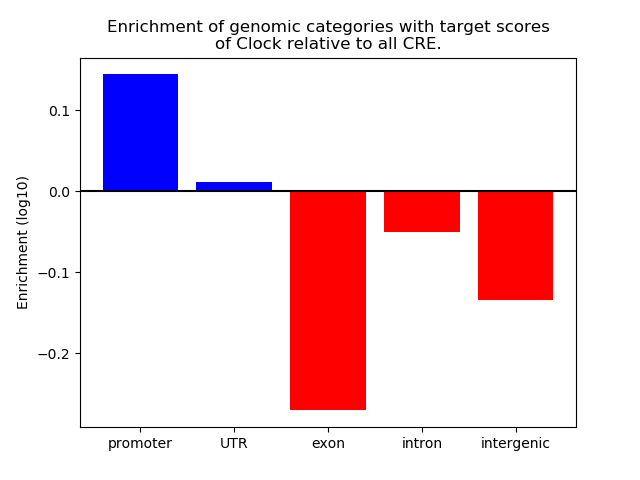
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_82845042_82846306 | 1.82 |
Rffl |
ring finger and FYVE like domain containing protein |
418 |
0.75 |
| chr3_96559570_96560186 | 1.59 |
Txnip |
thioredoxin interacting protein |
155 |
0.87 |
| chr13_5714127_5714791 | 1.46 |
Gm35330 |
predicted gene, 35330 |
9983 |
0.27 |
| chr4_154024404_154026596 | 1.43 |
Smim1 |
small integral membrane protein 1 |
116 |
0.93 |
| chr8_40863729_40864362 | 1.40 |
Slc7a2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
1467 |
0.34 |
| chr2_32602237_32602961 | 1.34 |
St6galnac6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
2873 |
0.11 |
| chr14_46761470_46761705 | 1.33 |
Cdkn3 |
cyclin-dependent kinase inhibitor 3 |
992 |
0.38 |
| chr3_14878521_14878851 | 1.27 |
Car2 |
carbonic anhydrase 2 |
7587 |
0.19 |
| chr4_46401911_46402304 | 1.19 |
Hemgn |
hemogen |
2129 |
0.22 |
| chr5_134913789_134913940 | 1.11 |
Cldn13 |
claudin 13 |
1662 |
0.18 |
| chr17_32402450_32402601 | 1.08 |
A530088E08Rik |
RIKEN cDNA A530088E08 gene |
481 |
0.59 |
| chr16_87705786_87706863 | 1.05 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
7320 |
0.23 |
| chr16_33755568_33756608 | 1.02 |
Heg1 |
heart development protein with EGF-like domains 1 |
435 |
0.83 |
| chr18_32557788_32558922 | 1.01 |
Gypc |
glycophorin C |
1625 |
0.41 |
| chr8_120368836_120369597 | 0.98 |
Gm22715 |
predicted gene, 22715 |
74333 |
0.08 |
| chr2_90667370_90668252 | 0.97 |
Nup160 |
nucleoporin 160 |
9404 |
0.2 |
| chr18_88924372_88924751 | 0.96 |
Socs6 |
suppressor of cytokine signaling 6 |
2920 |
0.21 |
| chr9_113990546_113990983 | 0.95 |
Fbxl2 |
F-box and leucine-rich repeat protein 2 |
740 |
0.65 |
| chr7_141009744_141010714 | 0.93 |
Ifitm3 |
interferon induced transmembrane protein 3 |
541 |
0.54 |
| chr12_98270951_98271119 | 0.91 |
Gpr65 |
G-protein coupled receptor 65 |
2400 |
0.21 |
| chr4_102570084_102571297 | 0.90 |
Pde4b |
phosphodiesterase 4B, cAMP specific |
595 |
0.85 |
| chr6_136809890_136810041 | 0.83 |
Gm44364 |
predicted gene, 44364 |
1282 |
0.22 |
| chr10_58674680_58675712 | 0.83 |
Edar |
ectodysplasin-A receptor |
458 |
0.8 |
| chr10_21668935_21669344 | 0.82 |
Gm5420 |
predicted gene 5420 |
17272 |
0.2 |
| chr10_89484249_89484400 | 0.81 |
Nr1h4 |
nuclear receptor subfamily 1, group H, member 4 |
22325 |
0.19 |
| chr19_8736753_8736904 | 0.81 |
Wdr74 |
WD repeat domain 74 |
684 |
0.3 |
| chr14_66241488_66242199 | 0.80 |
Ptk2b |
PTK2 protein tyrosine kinase 2 beta |
28291 |
0.16 |
| chr1_60852322_60852473 | 0.78 |
Gm38137 |
predicted gene, 38137 |
27941 |
0.11 |
| chr13_55756582_55757242 | 0.77 |
4930451E10Rik |
RIKEN cDNA 4930451E10 gene |
27567 |
0.1 |
| chrX_8271051_8272966 | 0.77 |
Slc38a5 |
solute carrier family 38, member 5 |
366 |
0.82 |
| chr19_46357434_46357942 | 0.77 |
Mfsd13a |
major facilitator superfamily domain containing 13a |
749 |
0.39 |
| chr16_32707369_32707640 | 0.75 |
Gm9556 |
predicted gene 9556 |
19162 |
0.12 |
| chr9_21029266_21030734 | 0.75 |
Icam4 |
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
546 |
0.46 |
| chr19_32322991_32323275 | 0.74 |
Sgms1 |
sphingomyelin synthase 1 |
96 |
0.98 |
| chr18_57085898_57086199 | 0.73 |
Gm18739 |
predicted gene, 18739 |
31232 |
0.15 |
| chr9_40765729_40765955 | 0.70 |
Gm48293 |
predicted gene, 48293 |
3169 |
0.13 |
| chr7_27447790_27448588 | 0.68 |
Blvrb |
biliverdin reductase B (flavin reductase (NADPH)) |
128 |
0.82 |
| chr2_103483073_103483738 | 0.63 |
Cat |
catalase |
1720 |
0.36 |
| chr12_53832808_53833508 | 0.62 |
1700060O08Rik |
RIKEN cDNA 1700060O08 gene |
246234 |
0.02 |
| chr9_108338333_108338531 | 0.61 |
Gm37401 |
predicted gene, 37401 |
159 |
0.75 |
| chr12_105026449_105026657 | 0.60 |
Gm47650 |
predicted gene, 47650 |
1410 |
0.24 |
| chr1_61402413_61403049 | 0.59 |
9530026F06Rik |
RIKEN cDNA 9530026F06 gene |
24299 |
0.16 |
| chr13_75709278_75709841 | 0.59 |
Ell2 |
elongation factor RNA polymerase II 2 |
1848 |
0.23 |
| chr12_33966605_33968831 | 0.59 |
Twist1 |
twist basic helix-loop-helix transcription factor 1 |
10047 |
0.22 |
| chr5_67821909_67822220 | 0.58 |
Atp8a1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
6211 |
0.14 |
| chr7_19675940_19676174 | 0.58 |
Apoc2 |
apolipoprotein C-II |
807 |
0.36 |
| chrX_143824621_143825251 | 0.58 |
Capn6 |
calpain 6 |
2396 |
0.29 |
| chr11_101316299_101317280 | 0.57 |
Psme3 |
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
188 |
0.83 |
| chr1_186358273_186358675 | 0.57 |
Gm37491 |
predicted gene, 37491 |
11159 |
0.28 |
| chr1_75217955_75219308 | 0.57 |
Tuba4a |
tubulin, alpha 4A |
7 |
0.92 |
| chr3_85991395_85991798 | 0.57 |
Prss48 |
protease, serine 48 |
10895 |
0.12 |
| chr8_33891636_33892433 | 0.57 |
Rbpms |
RNA binding protein gene with multiple splicing |
270 |
0.91 |
| chr18_56871689_56871916 | 0.56 |
Gm18087 |
predicted gene, 18087 |
44286 |
0.14 |
| chr10_127749619_127751660 | 0.56 |
Gpr182 |
G protein-coupled receptor 182 |
1093 |
0.28 |
| chr17_14249901_14250892 | 0.55 |
Gm34567 |
predicted gene, 34567 |
545 |
0.76 |
| chr13_118718500_118718874 | 0.55 |
Gm16263 |
predicted gene 16263 |
3291 |
0.24 |
| chr18_75502739_75503903 | 0.54 |
Gm10532 |
predicted gene 10532 |
11324 |
0.26 |
| chr16_8811666_8811841 | 0.53 |
1810013L24Rik |
RIKEN cDNA 1810013L24 gene |
18347 |
0.15 |
| chr15_37733527_37733723 | 0.53 |
Gm49397 |
predicted gene, 49397 |
710 |
0.42 |
| chr3_129331827_129332986 | 0.53 |
Enpep |
glutamyl aminopeptidase |
142 |
0.95 |
| chr14_20479030_20480298 | 0.52 |
Anxa7 |
annexin A7 |
244 |
0.88 |
| chr15_77928261_77928943 | 0.52 |
Txn2 |
thioredoxin 2 |
366 |
0.83 |
| chr11_89298118_89300658 | 0.51 |
Nog |
noggin |
2944 |
0.27 |
| chr2_132943595_132944461 | 0.51 |
Fermt1 |
fermitin family member 1 |
1601 |
0.32 |
| chr7_45474996_45475456 | 0.51 |
Dhdh |
dihydrodiol dehydrogenase (dimeric) |
4705 |
0.06 |
| chr5_119271318_119271616 | 0.50 |
n-R5s175 |
nuclear encoded rRNA 5S 175 |
25778 |
0.22 |
| chr11_86586191_86587628 | 0.49 |
Vmp1 |
vacuole membrane protein 1 |
28 |
0.97 |
| chr8_84810943_84811513 | 0.49 |
Nfix |
nuclear factor I/X |
10884 |
0.08 |
| chr7_49522042_49523415 | 0.49 |
Nav2 |
neuron navigator 2 |
25464 |
0.21 |
| chr8_105822112_105822566 | 0.49 |
Ranbp10 |
RAN binding protein 10 |
4866 |
0.1 |
| chr1_107529306_107529725 | 0.48 |
Serpinb10 |
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
488 |
0.8 |
| chr6_120579351_120580579 | 0.47 |
Gm44124 |
predicted gene, 44124 |
211 |
0.9 |
| chr6_127151640_127154010 | 0.47 |
Ccnd2 |
cyclin D2 |
632 |
0.57 |
| chr14_47747803_47748032 | 0.47 |
4930538L07Rik |
RIKEN cDNA 4930538L07 gene |
9569 |
0.16 |
| chr11_99016965_99017116 | 0.46 |
Gm23451 |
predicted gene, 23451 |
112 |
0.94 |
| chr18_37999322_37999473 | 0.46 |
Arap3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
428 |
0.71 |
| chr1_12875347_12875986 | 0.46 |
Sulf1 |
sulfatase 1 |
32875 |
0.18 |
| chr7_120835150_120835405 | 0.45 |
Eef2k |
eukaryotic elongation factor-2 kinase |
7554 |
0.14 |
| chr2_174110026_174111205 | 0.45 |
Npepl1 |
aminopeptidase-like 1 |
266 |
0.91 |
| chr6_5295172_5295334 | 0.45 |
Pon2 |
paraoxonase 2 |
3077 |
0.24 |
| chr9_67842828_67843201 | 0.44 |
Vps13c |
vacuolar protein sorting 13C |
2602 |
0.27 |
| chr6_119194389_119194967 | 0.44 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1553 |
0.37 |
| chr10_75568361_75569309 | 0.44 |
Ggt1 |
gamma-glutamyltransferase 1 |
161 |
0.91 |
| chr4_142020367_142020518 | 0.44 |
4930455G09Rik |
RIKEN cDNA 4930455G09 gene |
2544 |
0.19 |
| chr9_53402629_53402979 | 0.43 |
4930550C14Rik |
RIKEN cDNA 4930550C14 gene |
479 |
0.73 |
| chr14_27230459_27230927 | 0.43 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
7346 |
0.19 |
| chr2_118487225_118487434 | 0.42 |
Gm13983 |
predicted gene 13983 |
3152 |
0.2 |
| chr12_35534028_35535328 | 0.42 |
Ahr |
aryl-hydrocarbon receptor |
55 |
0.76 |
| chr16_44349131_44349726 | 0.41 |
Spice1 |
spindle and centriole associated protein 1 |
1382 |
0.45 |
| chr6_120179299_120179618 | 0.41 |
Ninj2 |
ninjurin 2 |
14365 |
0.19 |
| chr19_34290165_34291436 | 0.41 |
Fas |
Fas (TNF receptor superfamily member 6) |
118 |
0.96 |
| chr4_132342884_132343681 | 0.41 |
Rcc1 |
regulator of chromosome condensation 1 |
2075 |
0.11 |
| chr4_134759385_134759719 | 0.41 |
Ldlrap1 |
low density lipoprotein receptor adaptor protein 1 |
8472 |
0.2 |
| chr5_73629767_73629939 | 0.41 |
Lrrc66 |
leucine rich repeat containing 66 |
459 |
0.78 |
| chr17_5406629_5406997 | 0.40 |
Gm29050 |
predicted gene 29050 |
2692 |
0.23 |
| chr17_33916771_33917099 | 0.40 |
Tapbp |
TAP binding protein |
749 |
0.22 |
| chr7_28796055_28797204 | 0.40 |
Rinl |
Ras and Rab interactor-like |
309 |
0.76 |
| chr10_84550138_84550342 | 0.40 |
Platr7 |
pluripotency associated transcript 7 |
3098 |
0.16 |
| chr15_12545486_12545637 | 0.40 |
Pdzd2 |
PDZ domain containing 2 |
3717 |
0.23 |
| chr12_105036200_105036351 | 0.40 |
Glrx5 |
glutaredoxin 5 |
1059 |
0.33 |
| chr8_117201307_117201734 | 0.40 |
Gan |
giant axonal neuropathy |
43383 |
0.14 |
| chr8_84701273_84703379 | 0.38 |
Lyl1 |
lymphoblastomic leukemia 1 |
545 |
0.59 |
| chr14_30010042_30010685 | 0.38 |
Chdh |
choline dehydrogenase |
1192 |
0.28 |
| chr17_86437666_86438206 | 0.38 |
Prkce |
protein kinase C, epsilon |
52626 |
0.14 |
| chr17_29450623_29451950 | 0.38 |
Gm36199 |
predicted gene, 36199 |
2388 |
0.19 |
| chr2_163685701_163685852 | 0.37 |
Gm16316 |
predicted gene 16316 |
6503 |
0.15 |
| chr3_100480305_100480711 | 0.37 |
Tent5c |
terminal nucleotidyltransferase 5C |
8686 |
0.14 |
| chr17_88753020_88753753 | 0.37 |
Lhcgr |
luteinizing hormone/choriogonadotropin receptor |
20670 |
0.21 |
| chr4_119187058_119187758 | 0.37 |
Ermap |
erythroblast membrane-associated protein |
1339 |
0.24 |
| chr2_157202552_157202949 | 0.37 |
Rbl1 |
RB transcriptional corepressor like 1 |
1762 |
0.3 |
| chr3_152198770_152199020 | 0.36 |
Dnajb4 |
DnaJ heat shock protein family (Hsp40) member B4 |
5050 |
0.14 |
| chr16_93938522_93938959 | 0.36 |
Cldn14 |
claudin 14 |
3644 |
0.18 |
| chr12_17728003_17728647 | 0.36 |
Hpcal1 |
hippocalcin-like 1 |
99 |
0.98 |
| chr14_22289728_22289908 | 0.36 |
Lrmda |
leucine rich melanocyte differentiation associated |
20347 |
0.27 |
| chr18_34409435_34410714 | 0.36 |
Pkd2l2 |
polycystic kidney disease 2-like 2 |
648 |
0.7 |
| chr13_51652601_51652752 | 0.36 |
Secisbp2 |
SECIS binding protein 2 |
879 |
0.54 |
| chr1_9547698_9548725 | 0.35 |
Adhfe1 |
alcohol dehydrogenase, iron containing, 1 |
92 |
0.95 |
| chr12_56695635_56696302 | 0.35 |
Pax9 |
paired box 9 |
239 |
0.9 |
| chr5_135151111_135151379 | 0.35 |
Tbl2 |
transducin (beta)-like 2 |
1473 |
0.25 |
| chr13_47111734_47112164 | 0.35 |
1700026N04Rik |
RIKEN cDNA 1700026N04 gene |
5530 |
0.13 |
| chr5_124052007_124052188 | 0.34 |
Gm43661 |
predicted gene 43661 |
255 |
0.85 |
| chr11_30998188_30998421 | 0.34 |
Gm12102 |
predicted gene 12102 |
5594 |
0.18 |
| chr8_70329770_70331438 | 0.34 |
Gdf1 |
growth differentiation factor 1 |
787 |
0.45 |
| chr12_33218601_33218752 | 0.34 |
Atxn7l1os1 |
ataxin 7-like 1, opposite strand 1 |
10178 |
0.21 |
| chr1_193509891_193510142 | 0.34 |
Mir205hg |
Mir205 host gene |
158 |
0.94 |
| chr11_96790204_96790385 | 0.34 |
Cbx1 |
chromobox 1 |
1045 |
0.36 |
| chr17_35813042_35813313 | 0.34 |
Ier3 |
immediate early response 3 |
8507 |
0.07 |
| chr1_167269734_167270158 | 0.33 |
Uck2 |
uridine-cytidine kinase 2 |
14655 |
0.11 |
| chr4_116555353_116555929 | 0.33 |
Tmem69 |
transmembrane protein 69 |
294 |
0.81 |
| chr5_125056018_125058841 | 0.33 |
Gm42838 |
predicted gene 42838 |
412 |
0.71 |
| chr4_149696192_149696820 | 0.33 |
Pik3cd |
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
2046 |
0.23 |
| chr18_37742006_37744626 | 0.33 |
Pcdhgb6 |
protocadherin gamma subfamily B, 6 |
1222 |
0.15 |
| chr1_194943562_194943749 | 0.32 |
Cd34 |
CD34 antigen |
4660 |
0.15 |
| chr4_143141947_143142203 | 0.32 |
Mir7021 |
microRNA 7021 |
5913 |
0.16 |
| chr11_117962845_117962996 | 0.32 |
Socs3 |
suppressor of cytokine signaling 3 |
6266 |
0.15 |
| chr6_4509880_4510830 | 0.32 |
Gm43921 |
predicted gene, 43921 |
4268 |
0.17 |
| chr7_126862474_126862872 | 0.32 |
Hirip3 |
HIRA interacting protein 3 |
192 |
0.57 |
| chr2_119476817_119477902 | 0.32 |
Ino80 |
INO80 complex subunit |
254 |
0.91 |
| chr14_63164156_63165244 | 0.32 |
Fdft1 |
farnesyl diphosphate farnesyl transferase 1 |
225 |
0.91 |
| chr3_93558555_93558706 | 0.32 |
S100a10 |
S100 calcium binding protein A10 (calpactin) |
2097 |
0.21 |
| chr5_76947565_76947716 | 0.32 |
Ppat |
phosphoribosyl pyrophosphate amidotransferase |
3525 |
0.15 |
| chr18_61661645_61661818 | 0.32 |
Carmn |
cardiac mesoderm enhancer-associated non-coding RNA |
3759 |
0.13 |
| chr4_21849018_21849173 | 0.32 |
Pnisr |
PNN interacting serine/arginine-rich |
1014 |
0.55 |
| chr8_109185308_109186079 | 0.32 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
64173 |
0.13 |
| chr13_73499214_73499493 | 0.31 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
3508 |
0.26 |
| chr9_40487712_40488106 | 0.31 |
Gm5055 |
predicted gene 5055 |
189 |
0.93 |
| chr5_66079108_66079628 | 0.31 |
Rbm47 |
RNA binding motif protein 47 |
1616 |
0.27 |
| chr9_42461646_42462435 | 0.31 |
Tbcel |
tubulin folding cofactor E-like |
579 |
0.73 |
| chr19_44135200_44135854 | 0.30 |
Cwf19l1 |
CWF19-like 1, cell cycle control (S. pombe) |
98 |
0.95 |
| chr7_98175238_98175752 | 0.30 |
Gm16938 |
predicted gene, 16938 |
1699 |
0.27 |
| chr1_40324725_40326229 | 0.30 |
Il1rl2 |
interleukin 1 receptor-like 2 |
134 |
0.96 |
| chr4_63561098_63561515 | 0.30 |
Tmem268 |
transmembrane protein 268 |
946 |
0.42 |
| chr12_105962892_105963521 | 0.30 |
Vrk1 |
vaccinia related kinase 1 |
47022 |
0.13 |
| chr10_125321650_125321801 | 0.30 |
Slc16a7 |
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
6810 |
0.24 |
| chr7_49303593_49304723 | 0.30 |
Nav2 |
neuron navigator 2 |
20 |
0.98 |
| chr15_77977643_77977952 | 0.30 |
Eif3d |
eukaryotic translation initiation factor 3, subunit D |
6984 |
0.16 |
| chr10_42940424_42940575 | 0.30 |
Scml4 |
Scm polycomb group protein like 4 |
618 |
0.74 |
| chrX_140061291_140062848 | 0.30 |
Nup62cl |
nucleoporin 62 C-terminal like |
224 |
0.85 |
| chr1_52499555_52500406 | 0.29 |
Nab1 |
Ngfi-A binding protein 1 |
320 |
0.85 |
| chr1_129956479_129957256 | 0.29 |
Gm37278 |
predicted gene, 37278 |
13705 |
0.22 |
| chr13_21418981_21419424 | 0.28 |
Gm50481 |
predicted gene, 50481 |
7151 |
0.07 |
| chr17_46160338_46160791 | 0.28 |
Gtpbp2 |
GTP binding protein 2 |
468 |
0.68 |
| chr10_42273694_42273969 | 0.28 |
Foxo3 |
forkhead box O3 |
2865 |
0.34 |
| chr10_81319820_81320351 | 0.27 |
Cactin |
cactin, spliceosome C complex subunit |
1018 |
0.23 |
| chr8_84800470_84800683 | 0.27 |
Nfix |
nuclear factor I/X |
232 |
0.86 |
| chr11_98788809_98789966 | 0.27 |
Msl1 |
male specific lethal 1 |
6129 |
0.1 |
| chr13_104140168_104140319 | 0.27 |
Sgtb |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
30414 |
0.12 |
| chr16_76510697_76511196 | 0.27 |
Gm45030 |
predicted gene 45030 |
8037 |
0.24 |
| chr13_28770503_28770945 | 0.27 |
Gm17528 |
predicted gene, 17528 |
56399 |
0.12 |
| chr5_21046537_21046688 | 0.26 |
Ptpn12 |
protein tyrosine phosphatase, non-receptor type 12 |
9113 |
0.16 |
| chr9_88241670_88242042 | 0.26 |
Gm28231 |
predicted gene 28231 |
18039 |
0.16 |
| chr11_69751843_69752104 | 0.26 |
Polr2a |
polymerase (RNA) II (DNA directed) polypeptide A |
6250 |
0.06 |
| chr1_118310232_118310388 | 0.26 |
Tsn |
translin |
73 |
0.97 |
| chr11_29636734_29637040 | 0.26 |
Gm12092 |
predicted gene 12092 |
8622 |
0.14 |
| chr2_132110919_132111093 | 0.26 |
Slc23a2 |
solute carrier family 23 (nucleobase transporters), member 2 |
442 |
0.81 |
| chr1_75458926_75459599 | 0.26 |
Asic4 |
acid-sensing (proton-gated) ion channel family member 4 |
8563 |
0.08 |
| chr18_35553027_35554317 | 0.26 |
Snhg4 |
small nucleolar RNA host gene 4 |
234 |
0.75 |
| chr9_43204942_43206270 | 0.26 |
Pou2f3 |
POU domain, class 2, transcription factor 3 |
149 |
0.95 |
| chr5_103972357_103973054 | 0.26 |
Hsd17b13 |
hydroxysteroid (17-beta) dehydrogenase 13 |
4621 |
0.16 |
| chr11_32300067_32300964 | 0.26 |
Hbq1a |
hemoglobin, theta 1A |
446 |
0.71 |
| chr8_114641600_114641805 | 0.26 |
Gm16117 |
predicted gene 16117 |
412 |
0.87 |
| chr13_81931216_81931367 | 0.26 |
F830210D05Rik |
RIKEN cDNA F830210D05 gene |
1604 |
0.5 |
| chr2_73429039_73429190 | 0.26 |
Gm13707 |
predicted gene 13707 |
7205 |
0.14 |
| chr10_9699351_9699551 | 0.26 |
Gm6150 |
predicted gene 6150 |
14729 |
0.14 |
| chr16_18875243_18875394 | 0.25 |
Mrpl40 |
mitochondrial ribosomal protein L40 |
1221 |
0.32 |
| chr9_57439089_57439658 | 0.25 |
Ppcdc |
phosphopantothenoylcysteine decarboxylase |
718 |
0.55 |
| chr15_73059820_73059971 | 0.25 |
Trappc9 |
trafficking protein particle complex 9 |
1150 |
0.53 |
| chr5_111508721_111509927 | 0.25 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
72098 |
0.08 |
| chr13_6549550_6549952 | 0.25 |
Pitrm1 |
pitrilysin metallepetidase 1 |
1516 |
0.42 |
| chr8_47134944_47136227 | 0.25 |
E330018M18Rik |
RIKEN cDNA E330018M18 gene |
66 |
0.98 |
| chr17_66350075_66350634 | 0.25 |
Mtcl1 |
microtubule crosslinking factor 1 |
3809 |
0.22 |
| chr7_144940103_144940440 | 0.25 |
Ccnd1 |
cyclin D1 |
346 |
0.83 |
| chr10_107906593_107906899 | 0.25 |
Otogl |
otogelin-like |
5388 |
0.28 |
| chr7_131777798_131777949 | 0.25 |
Gm44547 |
predicted gene 44547 |
8706 |
0.22 |
| chr2_93447410_93447951 | 0.25 |
Cd82 |
CD82 antigen |
4999 |
0.18 |
| chr7_89521949_89522690 | 0.25 |
Prss23 |
protease, serine 23 |
3771 |
0.2 |
| chr2_11513131_11513381 | 0.24 |
Gm10851 |
predicted gene 10851 |
8450 |
0.14 |
| chr19_25675361_25675512 | 0.24 |
Dmrt2 |
doublesex and mab-3 related transcription factor 2 |
3025 |
0.26 |
| chr12_54405042_54405618 | 0.24 |
Gm46328 |
predicted gene, 46328 |
5154 |
0.16 |
| chr5_147825420_147826373 | 0.24 |
Gm43155 |
predicted gene 43155 |
3651 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 0.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 | 0.6 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.2 | 0.5 | GO:0046149 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.5 | GO:0015819 | lysine transport(GO:0015819) |
| 0.1 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.4 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.1 | 1.5 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 0.2 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.1 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.2 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.2 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.2 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 0.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.5 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.0 | 0.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.5 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0015744 | succinate transport(GO:0015744) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0036301 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0036491 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.2 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:2000665 | interleukin-5 secretion(GO:0072603) interleukin-13 secretion(GO:0072611) regulation of interleukin-5 secretion(GO:2000662) regulation of interleukin-13 secretion(GO:2000665) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.0 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.0 | GO:1901859 | negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.0 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.5 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.5 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:1901474 | L-histidine transmembrane transporter activity(GO:0005290) azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |