Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
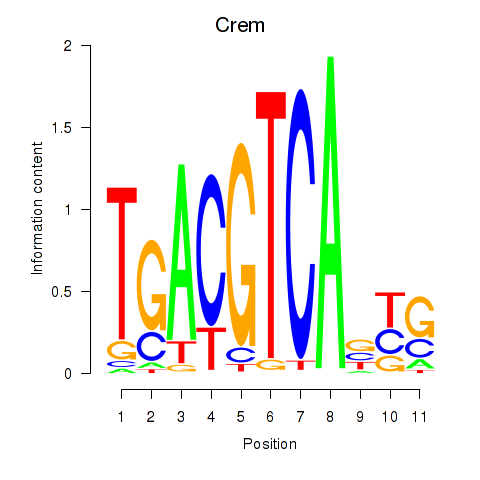
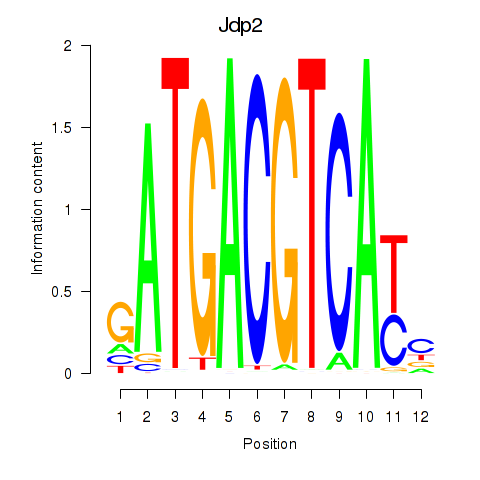
Results for Crem_Jdp2
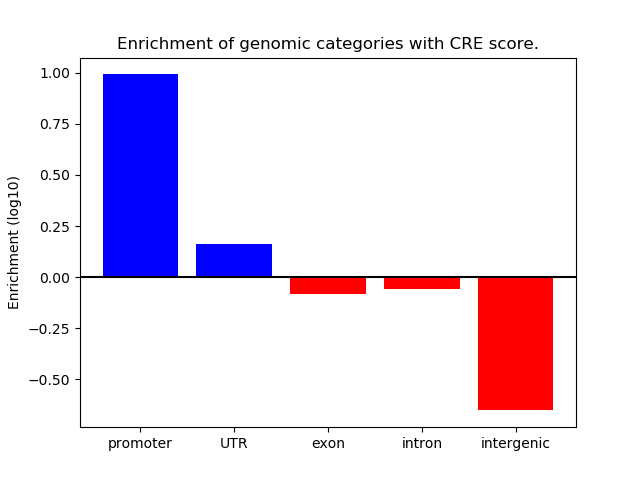
Z-value: 0.66
Transcription factors associated with Crem_Jdp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Crem
|
ENSMUSG00000063889.10 | cAMP responsive element modulator |
|
Jdp2
|
ENSMUSG00000034271.9 | Jun dimerization protein 2 |
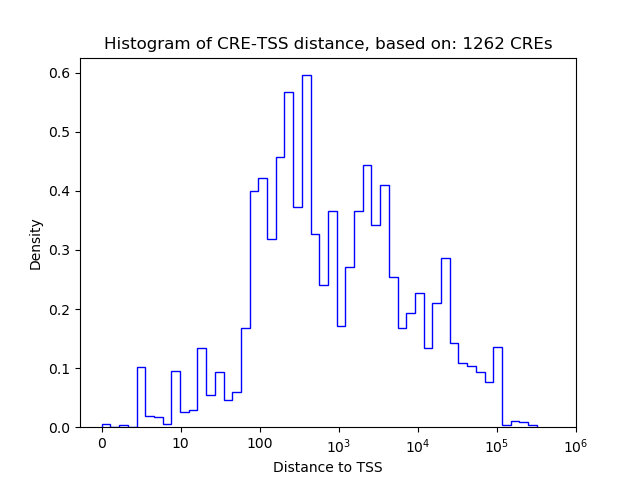
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_3281982_3282133 | Crem | 288 | 0.922724 | -0.40 | 1.6e-03 | Click! |
| chr18_3280402_3280691 | Crem | 532 | 0.811910 | -0.31 | 1.8e-02 | Click! |
| chr18_3281576_3281910 | Crem | 4 | 0.982127 | -0.25 | 5.3e-02 | Click! |
| chr18_3274616_3274842 | Crem | 6349 | 0.234001 | 0.12 | 3.5e-01 | Click! |
| chr18_3280827_3281518 | Crem | 40 | 0.980926 | -0.12 | 3.5e-01 | Click! |
| chr12_85601303_85602833 | Jdp2 | 154 | 0.942518 | -0.48 | 1.0e-04 | Click! |
| chr12_85581045_85581885 | Jdp2 | 17562 | 0.146422 | 0.34 | 7.4e-03 | Click! |
| chr12_85603052_85603203 | Jdp2 | 905 | 0.524648 | -0.33 | 9.3e-03 | Click! |
| chr12_85603259_85603447 | Jdp2 | 1131 | 0.437931 | -0.20 | 1.2e-01 | Click! |
| chr12_85571980_85572497 | Jdp2 | 26789 | 0.130692 | 0.16 | 2.2e-01 | Click! |
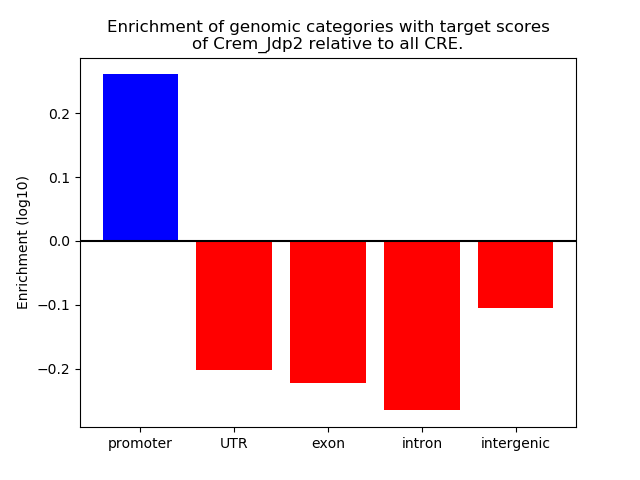
Activity of the Crem_Jdp2 motif across conditions
Conditions sorted by the z-value of the Crem_Jdp2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
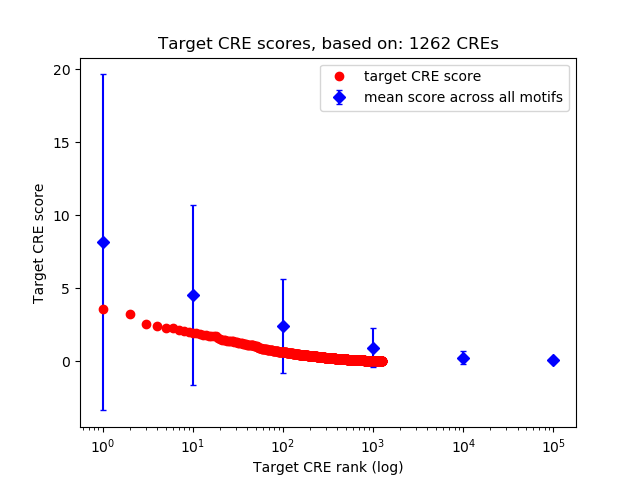
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_79498955_79500626 | 3.57 |
Mir9-3hg |
Mir9-3 host gene |
236 |
0.84 |
| chr1_79439702_79440415 | 3.24 |
Scg2 |
secretogranin II |
16 |
0.98 |
| chr18_35214570_35215443 | 2.56 |
Lrrtm2 |
leucine rich repeat transmembrane neuronal 2 |
2 |
0.52 |
| chr7_16982884_16983633 | 2.40 |
Gm42372 |
predicted gene, 42372 |
172 |
0.9 |
| chr11_115511512_115512231 | 2.30 |
Jpt1 |
Jupiter microtubule associated homolog 1 |
2245 |
0.14 |
| chr15_84104886_84106159 | 2.27 |
Sult4a1 |
sulfotransferase family 4A, member 1 |
87 |
0.93 |
| chr2_178141581_178143125 | 2.16 |
Phactr3 |
phosphatase and actin regulator 3 |
420 |
0.88 |
| chr3_17793835_17795104 | 2.08 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
427 |
0.75 |
| chr5_116589538_116590511 | 2.01 |
Srrm4 |
serine/arginine repetitive matrix 4 |
1793 |
0.34 |
| chr17_31809980_31810371 | 1.93 |
Gm49999 |
predicted gene, 49999 |
20649 |
0.15 |
| chr10_41072016_41072570 | 1.93 |
Gpr6 |
G protein-coupled receptor 6 |
8 |
0.97 |
| chrX_73063945_73064811 | 1.87 |
Pnma3 |
paraneoplastic antigen MA3 |
409 |
0.71 |
| chr5_117241784_117242411 | 1.83 |
Taok3 |
TAO kinase 3 |
1637 |
0.3 |
| chr11_6604576_6606131 | 1.81 |
Nacad |
NAC alpha domain containing |
700 |
0.46 |
| chr8_87539918_87540560 | 1.75 |
4933402J07Rik |
RIKEN cDNA 4933402J07 gene |
23614 |
0.14 |
| chr2_105668422_105670370 | 1.73 |
Pax6 |
paired box 6 |
461 |
0.65 |
| chr4_150651111_150652374 | 1.71 |
Slc45a1 |
solute carrier family 45, member 1 |
355 |
0.88 |
| chr9_91350199_91351559 | 1.70 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
74 |
0.95 |
| chr8_65617917_65619195 | 1.62 |
Marchf1 |
membrane associated ring-CH-type finger 1 |
31 |
0.99 |
| chr5_122636955_122637295 | 1.51 |
P2rx7 |
purinergic receptor P2X, ligand-gated ion channel, 7 |
6786 |
0.14 |
| chr5_104109155_104109917 | 1.48 |
Gm26703 |
predicted gene, 26703 |
245 |
0.88 |
| chr1_153660970_153662018 | 1.47 |
Rgs8 |
regulator of G-protein signaling 8 |
413 |
0.8 |
| chr15_25758674_25760120 | 1.45 |
Myo10 |
myosin X |
612 |
0.78 |
| chr18_76855816_76857376 | 1.40 |
Skor2 |
SKI family transcriptional corepressor 2 |
191 |
0.96 |
| chr3_34656299_34657721 | 1.39 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
974 |
0.4 |
| chr3_88214322_88216234 | 1.38 |
Mir3093 |
microRNA 3093 |
107 |
0.63 |
| chr12_111671006_111672264 | 1.38 |
Ckb |
creatine kinase, brain |
91 |
0.94 |
| chr13_83737592_83739114 | 1.36 |
Gm33366 |
predicted gene, 33366 |
182 |
0.66 |
| chr19_5084688_5085540 | 1.32 |
Tmem151a |
transmembrane protein 151A |
233 |
0.79 |
| chr18_28077967_28078215 | 1.29 |
Gm5064 |
predicted gene 5064 |
110726 |
0.07 |
| chr2_105673131_105674553 | 1.29 |
Pax6 |
paired box 6 |
121 |
0.95 |
| chr12_68997254_68997526 | 1.27 |
Gm47515 |
predicted gene, 47515 |
2420 |
0.27 |
| chr6_17748935_17749179 | 1.26 |
St7 |
suppression of tumorigenicity 7 |
113 |
0.56 |
| chr5_35454181_35454829 | 1.25 |
Gm43377 |
predicted gene 43377 |
58407 |
0.08 |
| chr10_25458395_25459149 | 1.24 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
8201 |
0.21 |
| chr2_94246278_94247531 | 1.20 |
Mir670hg |
MIR670 host gene (non-protein coding) |
3566 |
0.17 |
| chr3_94478073_94479450 | 1.20 |
Celf3 |
CUGBP, Elav-like family member 3 |
70 |
0.92 |
| chr15_98983254_98984205 | 1.19 |
4930578M01Rik |
RIKEN cDNA 4930578M01 gene |
102 |
0.93 |
| chr4_149854254_149854922 | 1.17 |
Gm47301 |
predicted gene, 47301 |
36824 |
0.09 |
| chr17_78454131_78454627 | 1.14 |
Gm19399 |
predicted gene, 19399 |
4266 |
0.19 |
| chr16_42444471_42445397 | 1.14 |
Gap43 |
growth associated protein 43 |
104283 |
0.07 |
| chr3_88831100_88832914 | 1.12 |
1500004A13Rik |
RIKEN cDNA 1500004A13 gene |
361 |
0.77 |
| chr8_12917045_12917331 | 1.12 |
Mcf2l |
mcf.2 transforming sequence-like |
1213 |
0.28 |
| chr7_79504311_79505700 | 1.12 |
Mir9-3 |
microRNA 9-3 |
259 |
0.82 |
| chr19_61226246_61226725 | 1.11 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
198 |
0.91 |
| chr17_56137995_56139079 | 1.09 |
Sema6b |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
1806 |
0.18 |
| chr3_88771097_88772528 | 1.08 |
Syt11 |
synaptotagmin XI |
787 |
0.45 |
| chr12_102554749_102554936 | 1.05 |
Chga |
chromogranin A |
127 |
0.96 |
| chr15_64922842_64923330 | 1.04 |
Adcy8 |
adenylate cyclase 8 |
790 |
0.73 |
| chr17_43952337_43954220 | 1.04 |
Rcan2 |
regulator of calcineurin 2 |
27 |
0.99 |
| chr18_31443497_31444446 | 1.03 |
Syt4 |
synaptotagmin IV |
3435 |
0.19 |
| chrX_66652653_66653969 | 0.99 |
Slitrk2 |
SLIT and NTRK-like family, member 2 |
304 |
0.9 |
| chr3_95224335_95224907 | 0.98 |
Mllt11 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
4056 |
0.09 |
| chr2_132781102_132782201 | 0.94 |
Chgb |
chromogranin B |
373 |
0.82 |
| chr6_83185720_83187846 | 0.92 |
Dctn1 |
dynactin 1 |
837 |
0.39 |
| chr8_54957303_54957776 | 0.91 |
Gm45263 |
predicted gene 45263 |
2280 |
0.24 |
| chr7_37768541_37768692 | 0.89 |
Zfp536 |
zinc finger protein 536 |
1008 |
0.62 |
| chr18_82825279_82826286 | 0.86 |
4930445N18Rik |
RIKEN cDNA 4930445N18 gene |
30112 |
0.12 |
| chr3_94483546_94484940 | 0.86 |
Celf3 |
CUGBP, Elav-like family member 3 |
84 |
0.93 |
| chr15_83509799_83509950 | 0.85 |
Ttll1 |
tubulin tyrosine ligase-like 1 |
919 |
0.44 |
| chr8_54956010_54956394 | 0.83 |
Gpm6a |
glycoprotein m6a |
1359 |
0.38 |
| chr15_69041723_69041971 | 0.82 |
Gm49422 |
predicted gene, 49422 |
82316 |
0.1 |
| chr8_70486586_70487634 | 0.82 |
Tmem59l |
transmembrane protein 59-like |
214 |
0.81 |
| chr9_87115556_87115707 | 0.82 |
Gm8282 |
predicted gene 8282 |
11703 |
0.17 |
| chr6_13837081_13837232 | 0.82 |
Gpr85 |
G protein-coupled receptor 85 |
85 |
0.97 |
| chr15_28181738_28182178 | 0.81 |
Gm49234 |
predicted gene, 49234 |
10164 |
0.17 |
| chr7_109780891_109781950 | 0.79 |
Nrip3 |
nuclear receptor interacting protein 3 |
123 |
0.94 |
| chr12_31711352_31711698 | 0.79 |
Gpr22 |
G protein-coupled receptor 22 |
2401 |
0.25 |
| chr6_124463804_124465512 | 0.78 |
Clstn3 |
calsyntenin 3 |
136 |
0.92 |
| chrX_153035799_153036204 | 0.77 |
Mageh1 |
melanoma antigen, family H, 1 |
1574 |
0.28 |
| chr1_79437543_79437779 | 0.77 |
Scg2 |
secretogranin II |
2381 |
0.37 |
| chr4_109976788_109977122 | 0.76 |
Dmrta2os |
doublesex and mab-3 related transcription factor like family A2, opposite strand |
536 |
0.69 |
| chr5_150906399_150906670 | 0.75 |
Gm43298 |
predicted gene 43298 |
23892 |
0.17 |
| chr7_63445472_63445728 | 0.75 |
Otud7a |
OTU domain containing 7A |
823 |
0.5 |
| chr5_150262108_150262988 | 0.74 |
Fry |
FRY microtubule binding protein |
2781 |
0.26 |
| chr19_61225550_61226044 | 0.74 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
307 |
0.84 |
| chr7_126949305_126949933 | 0.72 |
Asphd1 |
aspartate beta-hydroxylase domain containing 1 |
37 |
0.9 |
| chr5_39641102_39641253 | 0.71 |
Hs3st1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
3420 |
0.26 |
| chr9_58008709_58009443 | 0.71 |
Cyp11a1 |
cytochrome P450, family 11, subfamily a, polypeptide 1 |
2665 |
0.19 |
| chr9_40268795_40270233 | 0.71 |
Scn3b |
sodium channel, voltage-gated, type III, beta |
81 |
0.96 |
| chr13_23761268_23761510 | 0.71 |
H4c1 |
H4 clustered histone 1 |
159 |
0.77 |
| chr13_78181011_78182180 | 0.71 |
Gm38604 |
predicted gene, 38604 |
1564 |
0.29 |
| chr18_86473556_86473849 | 0.70 |
Gm50384 |
predicted gene, 50384 |
227 |
0.95 |
| chr7_62417581_62417734 | 0.70 |
Mkrn3 |
makorin, ring finger protein, 3 |
2482 |
0.24 |
| chr5_129225715_129226283 | 0.70 |
Rps16-ps2 |
ribosomal protein S16, pseudogene 2 |
97482 |
0.07 |
| chrX_143664015_143665455 | 0.69 |
Pak3 |
p21 (RAC1) activated kinase 3 |
156 |
0.98 |
| chr1_134079290_134079527 | 0.69 |
Btg2 |
BTG anti-proliferation factor 2 |
288 |
0.86 |
| chr3_3308823_3309359 | 0.68 |
Gm8747 |
predicted gene 8747 |
42064 |
0.2 |
| chr9_121403057_121404479 | 0.67 |
Trak1 |
trafficking protein, kinesin binding 1 |
290 |
0.91 |
| chr16_29837120_29837709 | 0.66 |
Gm32679 |
predicted gene, 32679 |
2686 |
0.29 |
| chr6_83038262_83039190 | 0.66 |
Loxl3 |
lysyl oxidase-like 3 |
326 |
0.68 |
| chr7_19932934_19933834 | 0.66 |
Igsf23 |
immunoglobulin superfamily, member 23 |
11514 |
0.07 |
| chr1_118434945_118435685 | 0.66 |
Clasp1 |
CLIP associating protein 1 |
15591 |
0.13 |
| chr17_24689022_24690262 | 0.65 |
Syngr3 |
synaptogyrin 3 |
313 |
0.69 |
| chr13_65258047_65259444 | 0.65 |
Gm10775 |
predicted gene 10775 |
794 |
0.38 |
| chr7_78882466_78883900 | 0.65 |
Mir7-2 |
microRNA 7-2 |
5094 |
0.13 |
| chr12_47164059_47165060 | 0.64 |
Gm36971 |
predicted gene, 36971 |
483 |
0.87 |
| chr13_48227672_48228587 | 0.64 |
Gm36346 |
predicted gene, 36346 |
9122 |
0.16 |
| chr15_84167436_84168909 | 0.63 |
Mir6392 |
microRNA 6392 |
98 |
0.77 |
| chr5_134102760_134102911 | 0.63 |
Castor2 |
cytosolic arginine sensor for mTORC1 subunit 2 |
2830 |
0.2 |
| chr12_103315511_103316208 | 0.63 |
Fam181a |
family with sequence similarity 181, member A |
900 |
0.36 |
| chr9_73967774_73969221 | 0.63 |
Unc13c |
unc-13 homolog C |
469 |
0.88 |
| chr1_132322108_132322259 | 0.63 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
5413 |
0.13 |
| chr18_37800729_37801183 | 0.62 |
Pcdhgc3 |
protocadherin gamma subfamily C, 3 |
5408 |
0.06 |
| chr3_89959021_89959708 | 0.62 |
Atp8b2 |
ATPase, class I, type 8B, member 2 |
190 |
0.88 |
| chr12_52009968_52010722 | 0.62 |
Dtd2 |
D-tyrosyl-tRNA deacylase 2 |
3844 |
0.19 |
| chr8_61227862_61229170 | 0.62 |
Sh3rf1 |
SH3 domain containing ring finger 1 |
4272 |
0.23 |
| chr14_52313293_52314443 | 0.62 |
Sall2 |
spalt like transcription factor 2 |
2455 |
0.13 |
| chr2_127520770_127522413 | 0.61 |
Kcnip3 |
Kv channel interacting protein 3, calsenilin |
221 |
0.89 |
| chr11_84042912_84043451 | 0.59 |
Synrg |
synergin, gamma |
16526 |
0.16 |
| chr9_119322832_119323536 | 0.59 |
Oxsr1 |
oxidative-stress responsive 1 |
757 |
0.52 |
| chr11_102466964_102467996 | 0.59 |
Itga2b |
integrin alpha 2b |
281 |
0.83 |
| chr7_121391567_121393161 | 0.58 |
Hs3st2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
74 |
0.93 |
| chr4_155344717_155346023 | 0.58 |
Prkcz |
protein kinase C, zeta |
12 |
0.96 |
| chr1_92522550_92522701 | 0.58 |
Olfr1414 |
olfactory receptor 1414 |
3964 |
0.13 |
| chr15_38373513_38374380 | 0.58 |
Gm41307 |
predicted gene, 41307 |
30208 |
0.13 |
| chrX_136290697_136291204 | 0.58 |
n-R5s12 |
nuclear encoded rRNA 5S 12 |
13445 |
0.09 |
| chr9_110051810_110053856 | 0.57 |
Map4 |
microtubule-associated protein 4 |
781 |
0.54 |
| chr2_25264308_25268001 | 0.56 |
Tprn |
taperin |
1410 |
0.14 |
| chr1_188714025_188714176 | 0.56 |
Gm25269 |
predicted gene, 25269 |
46376 |
0.18 |
| chr3_129536277_129537008 | 0.56 |
Gm43072 |
predicted gene 43072 |
1499 |
0.33 |
| chrX_170674573_170675954 | 0.55 |
Asmt |
acetylserotonin O-methyltransferase |
2619 |
0.41 |
| chr16_4790080_4790632 | 0.55 |
Cdip1 |
cell death inducing Trp53 target 1 |
64 |
0.96 |
| chr10_21144738_21145721 | 0.55 |
Gm26577 |
predicted gene, 26577 |
135 |
0.93 |
| chr2_116053626_116054503 | 0.55 |
Meis2 |
Meis homeobox 2 |
4384 |
0.2 |
| chr5_139324497_139325782 | 0.55 |
Adap1 |
ArfGAP with dual PH domains 1 |
483 |
0.72 |
| chr9_20976801_20976984 | 0.54 |
S1pr2 |
sphingosine-1-phosphate receptor 2 |
111 |
0.92 |
| chr5_134675713_134676967 | 0.54 |
Gm10369 |
predicted gene 10369 |
150 |
0.92 |
| chr12_4082205_4082840 | 0.53 |
Dnajc27 |
DnaJ heat shock protein family (Hsp40) member C27 |
61 |
0.97 |
| chr10_34316245_34317506 | 0.53 |
Nt5dc1 |
5'-nucleotidase domain containing 1 |
3192 |
0.16 |
| chr19_53340390_53340659 | 0.52 |
Mxi1 |
MAX interactor 1, dimerization protein |
10116 |
0.13 |
| chr2_147968209_147968363 | 0.52 |
9030622O22Rik |
RIKEN cDNA 9030622O22 gene |
7899 |
0.24 |
| chr6_142960447_142960598 | 0.51 |
St8sia1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
3619 |
0.18 |
| chr1_37299560_37299784 | 0.51 |
1700074A21Rik |
RIKEN cDNA 1700074A21 gene |
25 |
0.71 |
| chr9_112227323_112228073 | 0.50 |
2310075C17Rik |
RIKEN cDNA 2310075C17 gene |
119 |
0.96 |
| chr5_32713090_32713944 | 0.50 |
Gm43852 |
predicted gene 43852 |
471 |
0.7 |
| chr8_105606313_105606835 | 0.50 |
Ripor1 |
RHO family interacting cell polarization regulator 1 |
1319 |
0.23 |
| chr8_69625128_69625279 | 0.50 |
Zfp868 |
zinc finger protein 868 |
342 |
0.85 |
| chr10_128378554_128378705 | 0.49 |
Gm26308 |
predicted gene, 26308 |
730 |
0.32 |
| chr6_17583744_17584496 | 0.49 |
Met |
met proto-oncogene |
37147 |
0.15 |
| chr15_77245294_77246216 | 0.49 |
Rbfox2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
169 |
0.94 |
| chr8_69749413_69749661 | 0.48 |
Zfp963 |
zinc finger protein 963 |
425 |
0.75 |
| chr4_155893812_155895517 | 0.48 |
Acap3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2352 |
0.11 |
| chr8_70743042_70744602 | 0.48 |
Pde4c |
phosphodiesterase 4C, cAMP specific |
743 |
0.38 |
| chr15_78509793_78510375 | 0.47 |
Il2rb |
interleukin 2 receptor, beta chain |
14813 |
0.09 |
| chr1_165091570_165092283 | 0.47 |
4930568G15Rik |
RIKEN cDNA 4930568G15 gene |
2 |
0.98 |
| chr9_113811975_113812935 | 0.47 |
Clasp2 |
CLIP associating protein 2 |
131 |
0.97 |
| chr12_82469855_82470833 | 0.47 |
Gm5435 |
predicted gene 5435 |
26193 |
0.18 |
| chr18_31316134_31317386 | 0.47 |
Rit2 |
Ras-like without CAAX 2 |
351 |
0.9 |
| chr2_180892979_180894605 | 0.47 |
Mir124a-3 |
microRNA 124a-3 |
248 |
0.53 |
| chr2_118662893_118663233 | 0.46 |
Pak6 |
p21 (RAC1) activated kinase 6 |
240 |
0.9 |
| chr15_72806569_72807700 | 0.46 |
Peg13 |
paternally expressed 13 |
3190 |
0.32 |
| chr3_4798346_4798833 | 0.46 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
119 |
0.97 |
| chr3_132880896_132881303 | 0.45 |
Gm29811 |
predicted gene, 29811 |
2503 |
0.24 |
| chr11_94043944_94044915 | 0.45 |
Spag9 |
sperm associated antigen 9 |
64 |
0.97 |
| chr6_91838887_91839258 | 0.45 |
Grip2 |
glutamate receptor interacting protein 2 |
11822 |
0.16 |
| chr19_25622238_25623053 | 0.44 |
Dmrt3 |
doublesex and mab-3 related transcription factor 3 |
12108 |
0.21 |
| chr16_90709814_90710620 | 0.44 |
Mis18a |
MIS18 kinetochore protein A |
16922 |
0.13 |
| chr19_6501022_6501221 | 0.44 |
Nrxn2 |
neurexin II |
3286 |
0.16 |
| chr16_92340779_92341614 | 0.44 |
Smim34 |
small integral membrane protein 34 |
11942 |
0.11 |
| chr16_87128194_87128870 | 0.44 |
Gm32865 |
predicted gene, 32865 |
93 |
0.98 |
| chr13_100892366_100893024 | 0.44 |
Gm41041 |
predicted gene, 41041 |
17194 |
0.14 |
| chr15_91017138_91018295 | 0.44 |
Kif21a |
kinesin family member 21A |
32102 |
0.16 |
| chr12_24829454_24829782 | 0.44 |
Mboat2 |
membrane bound O-acyltransferase domain containing 2 |
1261 |
0.32 |
| chr7_4149072_4149804 | 0.44 |
Leng9 |
leukocyte receptor cluster (LRC) member 9 |
936 |
0.34 |
| chr4_127549306_127549951 | 0.43 |
A630031M04Rik |
RIKEN cDNA A630031M04 gene |
4527 |
0.31 |
| chr7_118583609_118584872 | 0.43 |
Tmc7 |
transmembrane channel-like gene family 7 |
446 |
0.77 |
| chr18_6512631_6512915 | 0.43 |
Epc1 |
enhancer of polycomb homolog 1 |
3335 |
0.21 |
| chr5_147399872_147401256 | 0.42 |
Flt3 |
FMS-like tyrosine kinase 3 |
75 |
0.91 |
| chr3_116222523_116223160 | 0.42 |
Gm31651 |
predicted gene, 31651 |
22433 |
0.15 |
| chr1_165999821_165999972 | 0.42 |
Pou2f1 |
POU domain, class 2, transcription factor 1 |
2713 |
0.21 |
| chr1_6734529_6735444 | 0.42 |
St18 |
suppression of tumorigenicity 18 |
116 |
0.98 |
| chr17_90451511_90451662 | 0.42 |
Nrxn1 |
neurexin I |
3236 |
0.28 |
| chr9_57467083_57468096 | 0.42 |
Scamp5 |
secretory carrier membrane protein 5 |
406 |
0.73 |
| chr13_99493038_99493247 | 0.42 |
5330431K02Rik |
RIKEN cDNA 5330431K02 gene |
11297 |
0.16 |
| chr5_25222603_25223750 | 0.42 |
E130116L18Rik |
RIKEN cDNA E130116L18 gene |
23 |
0.81 |
| chr11_114793777_114794456 | 0.41 |
Btbd17 |
BTB (POZ) domain containing 17 |
1757 |
0.26 |
| chr7_48963141_48964311 | 0.41 |
Nav2 |
neuron navigator 2 |
4629 |
0.19 |
| chr8_31113321_31113667 | 0.41 |
Rnf122 |
ring finger protein 122 |
1630 |
0.32 |
| chr2_19909579_19910504 | 0.41 |
Etl4 |
enhancer trap locus 4 |
261 |
0.91 |
| chr10_80300884_80302968 | 0.41 |
Apc2 |
APC regulator of WNT signaling pathway 2 |
106 |
0.9 |
| chr9_108824114_108825614 | 0.41 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr5_69341722_69341873 | 0.41 |
Kctd8 |
potassium channel tetramerisation domain containing 8 |
118 |
0.92 |
| chr14_55560478_55560708 | 0.41 |
Dcaf11 |
DDB1 and CUL4 associated factor 11 |
59 |
0.64 |
| chr9_45054509_45055372 | 0.40 |
Mpzl3 |
myelin protein zero-like 3 |
246 |
0.61 |
| chr9_108787261_108787546 | 0.40 |
Ip6k2 |
inositol hexaphosphate kinase 2 |
3538 |
0.12 |
| chr13_20939238_20939491 | 0.40 |
Aoah |
acyloxyacyl hydrolase |
71269 |
0.09 |
| chr10_115586791_115588064 | 0.40 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
66 |
0.97 |
| chr9_4793004_4793465 | 0.40 |
Gria4 |
glutamate receptor, ionotropic, AMPA4 (alpha 4) |
2285 |
0.46 |
| chr18_83192582_83193531 | 0.40 |
1700095A13Rik |
RIKEN cDNA 1700095A13 gene |
85166 |
0.08 |
| chr6_28980713_28981850 | 0.40 |
Gm3294 |
predicted gene 3294 |
353 |
0.88 |
| chr13_44329334_44329485 | 0.39 |
Gm29676 |
predicted gene, 29676 |
48960 |
0.12 |
| chr6_127119580_127119731 | 0.39 |
Gm43126 |
predicted gene 43126 |
3311 |
0.14 |
| chr4_41568273_41569615 | 0.39 |
Fam219a |
family with sequence similarity 219, member A |
565 |
0.5 |
| chr15_81926465_81926674 | 0.39 |
Polr3h |
polymerase (RNA) III (DNA directed) polypeptide H |
329 |
0.77 |
| chr16_16828695_16829635 | 0.38 |
Spag6l |
sperm associated antigen 6-like |
116 |
0.93 |
| chr8_123410787_123412789 | 0.38 |
Tubb3 |
tubulin, beta 3 class III |
198 |
0.84 |
| chr11_52764429_52765649 | 0.38 |
Fstl4 |
follistatin-like 4 |
405 |
0.91 |
| chr15_81936444_81938042 | 0.38 |
Csdc2 |
cold shock domain containing C2, RNA binding |
261 |
0.82 |
| chr3_98095149_98095420 | 0.37 |
Gm42820 |
predicted gene 42820 |
26304 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 3.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 2.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 2.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 0.5 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.2 | 1.2 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.2 | 1.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 1.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.5 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.6 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 | 1.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.4 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 1.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 1.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.7 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.2 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.6 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.1 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.2 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 0.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.1 | 0.2 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 1.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.5 | GO:1902855 | regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.0 | 0.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1903288 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.0 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 | 0.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 1.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.0 | 0.6 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0042321 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0061205 | paramesonephric duct development(GO:0061205) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.3 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.0 | GO:2000822 | regulation of fear response(GO:1903365) positive regulation of fear response(GO:1903367) regulation of behavioral fear response(GO:2000822) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 3.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 2.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 2.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 0.8 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 3.0 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.1 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 2.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0034871 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.7 | GO:0052634 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0010435 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate 2-phosphatase activity(GO:0004083) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 1.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |