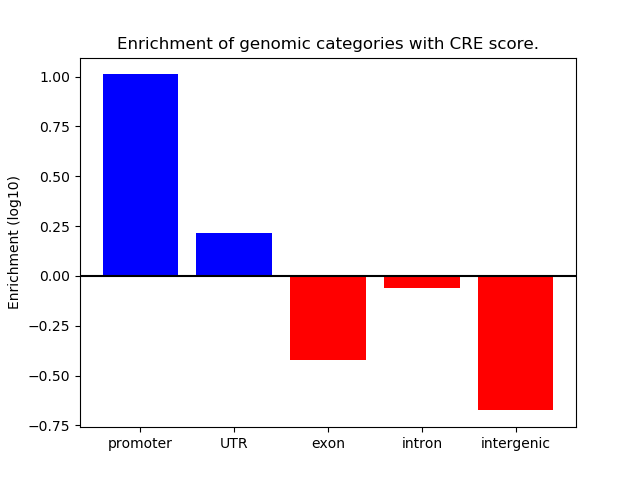
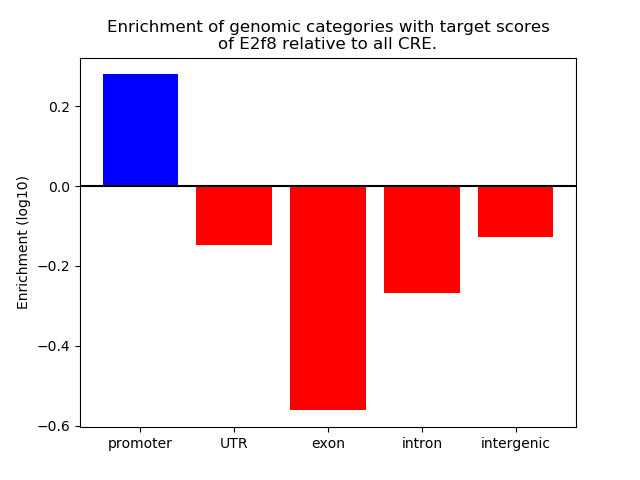
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
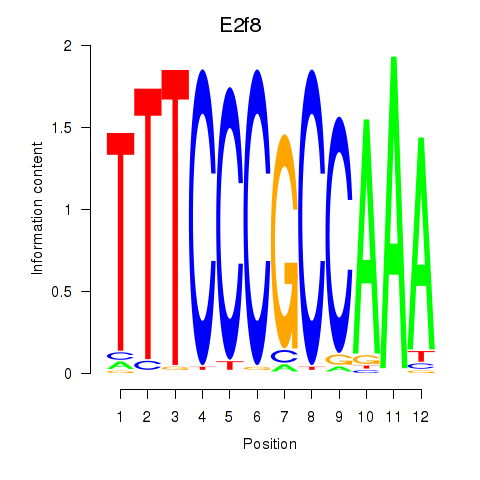
Results for E2f8
Z-value: 0.44
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSMUSG00000046179.11 | E2F transcription factor 8 |
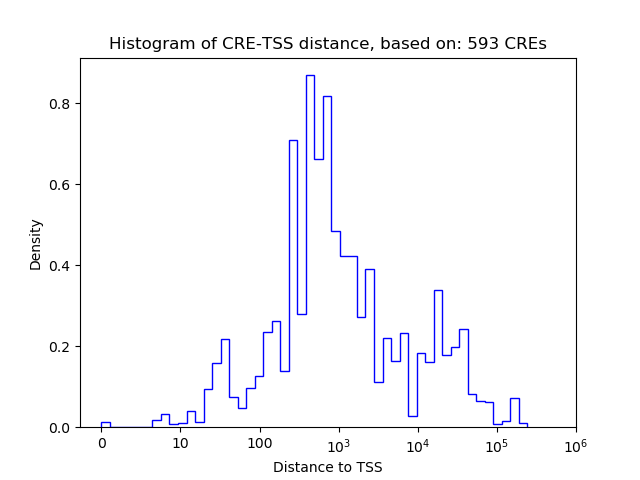
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_48881832_48882357 | E2f8 | 498 | 0.562900 | 0.59 | 5.8e-07 | Click! |
| chr7_48880272_48881025 | E2f8 | 356 | 0.649135 | 0.55 | 6.5e-06 | Click! |
| chr7_48879180_48879915 | E2f8 | 1457 | 0.262861 | 0.50 | 5.5e-05 | Click! |
| chr7_48881221_48881734 | E2f8 | 119 | 0.577878 | 0.34 | 8.2e-03 | Click! |
| chr7_48865527_48865678 | E2f8 | 9649 | 0.132537 | -0.26 | 4.1e-02 | Click! |
Activity of the E2f8 motif across conditions
Conditions sorted by the z-value of the E2f8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
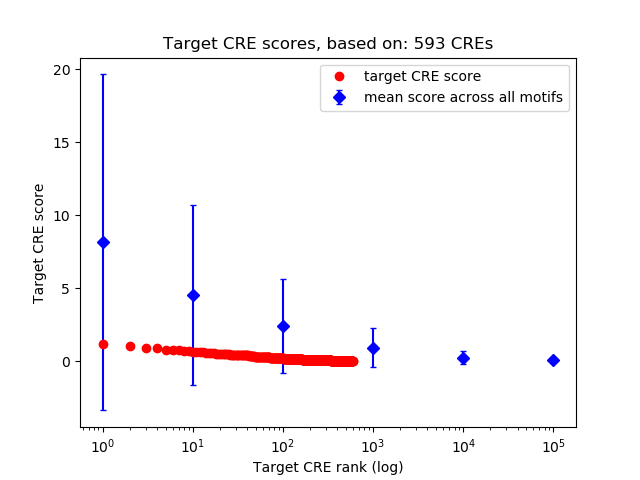
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_3398628_3399056 | 1.15 |
Samd9l |
sterile alpha motif domain containing 9-like |
730 |
0.64 |
| chr12_24708730_24709261 | 1.08 |
Rrm2 |
ribonucleotide reductase M2 |
34 |
0.97 |
| chr11_117810234_117810545 | 0.89 |
Syngr2 |
synaptogyrin 2 |
677 |
0.33 |
| chr11_98907694_98909099 | 0.88 |
Cdc6 |
cell division cycle 6 |
245 |
0.86 |
| chr6_127034730_127034972 | 0.78 |
Fgf6 |
fibroblast growth factor 6 |
19265 |
0.11 |
| chr5_17848306_17848609 | 0.78 |
Cd36 |
CD36 molecule |
1272 |
0.61 |
| chr19_6305921_6306134 | 0.76 |
Cdc42bpg |
CDC42 binding protein kinase gamma (DMPK-like) |
429 |
0.64 |
| chr13_51648729_51649046 | 0.73 |
Gm22806 |
predicted gene, 22806 |
688 |
0.62 |
| chr9_100643671_100643909 | 0.71 |
Stag1 |
stromal antigen 1 |
20 |
0.97 |
| chr8_23033869_23034540 | 0.65 |
Ank1 |
ankyrin 1, erythroid |
895 |
0.59 |
| chr2_25195300_25197035 | 0.64 |
Tor4a |
torsin family 4, member A |
592 |
0.43 |
| chr18_39485704_39487567 | 0.61 |
Nr3c1 |
nuclear receptor subfamily 3, group C, member 1 |
597 |
0.82 |
| chr16_35906184_35906335 | 0.60 |
Gm10237 |
predicted gene 10237 |
14205 |
0.1 |
| chr6_5154616_5154962 | 0.60 |
Pon1 |
paraoxonase 1 |
38974 |
0.14 |
| chr4_3937146_3938329 | 0.60 |
Plag1 |
pleiomorphic adenoma gene 1 |
645 |
0.54 |
| chr15_55090547_55090998 | 0.54 |
Dscc1 |
DNA replication and sister chromatid cohesion 1 |
281 |
0.87 |
| chr8_109737737_109738073 | 0.54 |
Atxn1l |
ataxin 1-like |
166 |
0.94 |
| chr14_31020219_31020404 | 0.52 |
Pbrm1 |
polybromo 1 |
1103 |
0.23 |
| chr11_29172094_29172676 | 0.51 |
Ppp4r3b |
protein phosphatase 4 regulatory subunit 3B |
505 |
0.68 |
| chr10_81140191_81141251 | 0.49 |
Zbtb7a |
zinc finger and BTB domain containing 7a |
2768 |
0.1 |
| chr5_42777354_42777505 | 0.49 |
Gm5554 |
predicted gene 5554 |
187180 |
0.03 |
| chr16_17619128_17619354 | 0.48 |
Smpd4 |
sphingomyelin phosphodiesterase 4 |
113 |
0.94 |
| chr5_77405086_77405486 | 0.48 |
Igfbp7 |
insulin-like growth factor binding protein 7 |
2754 |
0.21 |
| chr15_84719703_84721036 | 0.47 |
Gm20556 |
predicted gene, 20556 |
103 |
0.74 |
| chrX_101527905_101528056 | 0.47 |
Taf1 |
TATA-box binding protein associated factor 1 |
4754 |
0.17 |
| chr16_16146027_16146765 | 0.46 |
Spidr |
scaffolding protein involved in DNA repair |
408 |
0.87 |
| chrX_105069174_105070104 | 0.46 |
5530601H04Rik |
RIKEN cDNA 5530601H04 gene |
396 |
0.82 |
| chr17_56040358_56041583 | 0.44 |
Chaf1a |
chromatin assembly factor 1, subunit A (p150) |
531 |
0.58 |
| chr13_52584896_52585203 | 0.44 |
Syk |
spleen tyrosine kinase |
1546 |
0.51 |
| chrX_103622717_103623702 | 0.44 |
Ftx |
Ftx transcript, Xist regulator (non-protein coding) |
454 |
0.67 |
| chr7_67917220_67917371 | 0.44 |
Gm44666 |
predicted gene 44666 |
30710 |
0.15 |
| chr12_24706703_24706854 | 0.43 |
Rrm2 |
ribonucleotide reductase M2 |
1463 |
0.32 |
| chr11_90388814_90389066 | 0.43 |
Hlf |
hepatic leukemia factor |
650 |
0.8 |
| chrX_167228790_167228941 | 0.42 |
Tmsb4x |
thymosin, beta 4, X chromosome |
19550 |
0.17 |
| chr18_42513548_42513699 | 0.42 |
Tcerg1 |
transcription elongation regulator 1 (CA150) |
2100 |
0.3 |
| chrX_169418936_169419087 | 0.42 |
Gm15245 |
predicted gene 15245 |
28819 |
0.19 |
| chr9_51009335_51009739 | 0.41 |
Sik2 |
salt inducible kinase 2 |
464 |
0.81 |
| chr3_126996871_126998855 | 0.41 |
Ank2 |
ankyrin 2, brain |
575 |
0.63 |
| chr19_4155059_4155512 | 0.41 |
Ptprcap |
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
679 |
0.33 |
| chr16_91417240_91417391 | 0.41 |
Il10rb |
interleukin 10 receptor, beta |
10871 |
0.1 |
| chr19_24279590_24279827 | 0.40 |
Fxn |
frataxin |
860 |
0.58 |
| chr2_61655339_61656639 | 0.39 |
Tank |
TRAF family member-associated Nf-kappa B activator |
12158 |
0.22 |
| chr10_111010981_111011344 | 0.38 |
Zdhhc17 |
zinc finger, DHHC domain containing 17 |
1022 |
0.57 |
| chr12_55835950_55836196 | 0.38 |
Brms1l |
breast cancer metastasis-suppressor 1-like |
251 |
0.89 |
| chr6_29495887_29496782 | 0.35 |
Kcp |
kielin/chordin-like protein |
420 |
0.71 |
| chr1_132138543_132139629 | 0.34 |
Gm29629 |
predicted gene 29629 |
418 |
0.51 |
| chr8_94917668_94918304 | 0.34 |
Ccdc102a |
coiled-coil domain containing 102A |
112 |
0.94 |
| chr15_78845284_78846270 | 0.33 |
Cdc42ep1 |
CDC42 effector protein (Rho GTPase binding) 1 |
3153 |
0.12 |
| chr9_57763587_57763764 | 0.33 |
Clk3 |
CDC-like kinase 3 |
633 |
0.65 |
| chr6_34780141_34780388 | 0.32 |
Agbl3 |
ATP/GTP binding protein-like 3 |
168 |
0.92 |
| chr7_19769512_19770898 | 0.32 |
Bcam |
basal cell adhesion molecule |
290 |
0.54 |
| chr1_77226872_77227510 | 0.32 |
Gm38265 |
predicted gene, 38265 |
80199 |
0.1 |
| chr12_4233425_4233915 | 0.30 |
Cenpo |
centromere protein O |
268 |
0.55 |
| chr13_4942359_4942524 | 0.30 |
Gm47450 |
predicted gene, 47450 |
611 |
0.84 |
| chr17_71259969_71260163 | 0.30 |
Emilin2 |
elastin microfibril interfacer 2 |
3720 |
0.19 |
| chr12_102757182_102757722 | 0.29 |
Gm28051 |
predicted gene, 28051 |
313 |
0.38 |
| chr8_46615820_46616038 | 0.29 |
Primpol |
primase and polymerase (DNA-directed) |
1253 |
0.31 |
| chr17_84878392_84878652 | 0.28 |
Gm49982 |
predicted gene, 49982 |
24047 |
0.14 |
| chr7_5062843_5063444 | 0.28 |
U2af2 |
U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 2 |
839 |
0.26 |
| chr2_128157307_128157458 | 0.28 |
Bcl2l11 |
BCL2-like 11 (apoptosis facilitator) |
29251 |
0.16 |
| chr15_8967409_8968940 | 0.27 |
Ranbp3l |
RAN binding protein 3-like |
172 |
0.96 |
| chr12_56529878_56530592 | 0.27 |
Sfta3-ps |
surfactant associated 3, pseudogene |
808 |
0.49 |
| chr12_12941909_12942444 | 0.27 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
262 |
0.88 |
| chrX_155337145_155337602 | 0.27 |
Prdx4 |
peroxiredoxin 4 |
1094 |
0.5 |
| chr3_103777430_103777672 | 0.27 |
Gm15886 |
predicted gene 15886 |
136 |
0.9 |
| chr18_35739288_35740804 | 0.26 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
30 |
0.95 |
| chr5_139406928_139407938 | 0.26 |
C130050O18Rik |
RIKEN cDNA C130050O18 gene |
968 |
0.41 |
| chr4_115062473_115064166 | 0.26 |
Tal1 |
T cell acute lymphocytic leukemia 1 |
3811 |
0.18 |
| chr10_83353735_83354451 | 0.26 |
D10Wsu102e |
DNA segment, Chr 10, Wayne State University 102, expressed |
6128 |
0.17 |
| chrX_85576113_85576674 | 0.26 |
Tab3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
2243 |
0.33 |
| chr9_115165880_115166031 | 0.26 |
Gm4665 |
predicted gene 4665 |
20103 |
0.15 |
| chr8_112010916_112011253 | 0.26 |
Kars |
lysyl-tRNA synthetase |
182 |
0.65 |
| chr8_41055977_41056328 | 0.26 |
Mtus1 |
mitochondrial tumor suppressor 1 |
1358 |
0.36 |
| chr13_23695605_23696868 | 0.26 |
H1f6 |
H1.6 linker histone, cluster member |
422 |
0.53 |
| chr8_48921826_48922154 | 0.25 |
Gm25830 |
predicted gene, 25830 |
36491 |
0.18 |
| chr15_36779468_36779639 | 0.25 |
Ywhaz |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
12861 |
0.15 |
| chr2_103117112_103117263 | 0.25 |
Gm10038 |
predicted gene 10038 |
18032 |
0.16 |
| chr2_130663815_130664360 | 0.25 |
Ddrgk1 |
DDRGK domain containing 1 |
78 |
0.95 |
| chr18_38542410_38543000 | 0.24 |
Gm19100 |
predicted gene, 19100 |
40145 |
0.12 |
| chr3_95661677_95663214 | 0.24 |
Mcl1 |
myeloid cell leukemia sequence 1 |
3588 |
0.13 |
| chr11_26386745_26387188 | 0.24 |
Fancl |
Fanconi anemia, complementation group L |
236 |
0.94 |
| chr13_30338441_30338963 | 0.23 |
Agtr1a |
angiotensin II receptor, type 1a |
2258 |
0.33 |
| chr1_128591274_128591937 | 0.23 |
Cxcr4 |
chemokine (C-X-C motif) receptor 4 |
685 |
0.72 |
| chr11_88194059_88194740 | 0.23 |
Cuedc1 |
CUE domain containing 1 |
6578 |
0.18 |
| chr11_103131831_103132857 | 0.22 |
Hexim2 |
hexamethylene bis-acetamide inducible 2 |
85 |
0.95 |
| chr2_93460789_93461036 | 0.22 |
Cd82 |
CD82 antigen |
1000 |
0.52 |
| chr5_31494777_31495631 | 0.22 |
4930566F21Rik |
RIKEN cDNA 4930566F21 gene |
25 |
0.71 |
| chrX_96095788_96097365 | 0.22 |
Msn |
moesin |
479 |
0.84 |
| chr1_86581881_86582496 | 0.22 |
Pde6d |
phosphodiesterase 6D, cGMP-specific, rod, delta |
329 |
0.68 |
| chr18_74216226_74217021 | 0.22 |
Cxxc1 |
CXXC finger 1 (PHD domain) |
492 |
0.76 |
| chr7_36166793_36167655 | 0.21 |
Rpl17-ps9 |
ribosomal protein L17, pseudogene 9 |
48950 |
0.14 |
| chr11_70653066_70653217 | 0.21 |
Pfn1 |
profilin 1 |
1457 |
0.15 |
| chr1_173333573_173334024 | 0.21 |
Ackr1 |
atypical chemokine receptor 1 (Duffy blood group) |
48 |
0.97 |
| chr12_109454708_109455197 | 0.21 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
757 |
0.48 |
| chr16_22263818_22263981 | 0.21 |
Tra2b |
transformer 2 beta |
2001 |
0.22 |
| chr7_16048455_16049177 | 0.20 |
Bicra |
BRD4 interacting chromatin remodeling complex associated protein |
895 |
0.48 |
| chr11_115133328_115133870 | 0.20 |
Cd300lf |
CD300 molecule like family member F |
392 |
0.78 |
| chr3_130708012_130708163 | 0.20 |
Ostc |
oligosaccharyltransferase complex subunit (non-catalytic) |
1329 |
0.3 |
| chr5_119669544_119672401 | 0.20 |
Tbx3 |
T-box 3 |
46 |
0.85 |
| chr2_164769936_164770914 | 0.20 |
Ube2c |
ubiquitin-conjugating enzyme E2C |
522 |
0.43 |
| chr2_141701214_141701365 | 0.20 |
Gm25583 |
predicted gene, 25583 |
60799 |
0.15 |
| chr10_4335326_4335935 | 0.19 |
Akap12 |
A kinase (PRKA) anchor protein (gravin) 12 |
1085 |
0.51 |
| chr16_32163732_32163961 | 0.19 |
Nrros |
negative regulator of reactive oxygen species |
1611 |
0.27 |
| chr5_115011202_115012397 | 0.18 |
Sppl3 |
signal peptide peptidase 3 |
302 |
0.81 |
| chr15_82340829_82341556 | 0.18 |
Pheta2 |
PH domain containing endocytic trafficking adaptor 2 |
13 |
0.7 |
| chr9_96713030_96713604 | 0.18 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
6248 |
0.17 |
| chr3_41522196_41522994 | 0.18 |
2400006E01Rik |
RIKEN cDNA 2400006E01 gene |
622 |
0.65 |
| chr2_26504256_26504407 | 0.18 |
Notch1 |
notch 1 |
507 |
0.67 |
| chr17_31854730_31855751 | 0.17 |
Sik1 |
salt inducible kinase 1 |
553 |
0.73 |
| chr19_10060014_10060673 | 0.17 |
Fads3 |
fatty acid desaturase 3 |
4091 |
0.15 |
| chr13_5863142_5863416 | 0.17 |
Klf6 |
Kruppel-like factor 6 |
1399 |
0.37 |
| chr16_35938449_35939599 | 0.17 |
Parp9 |
poly (ADP-ribose) polymerase family, member 9 |
6 |
0.62 |
| chr11_98446534_98448432 | 0.17 |
Grb7 |
growth factor receptor bound protein 7 |
415 |
0.68 |
| chr6_144779851_144780433 | 0.16 |
Sox5 |
SRY (sex determining region Y)-box 5 |
1777 |
0.35 |
| chr5_116023370_116024380 | 0.16 |
Prkab1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
481 |
0.72 |
| chr2_165503533_165504813 | 0.16 |
Slc2a10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
386 |
0.83 |
| chr7_83655450_83655601 | 0.16 |
Il16 |
interleukin 16 |
30 |
0.96 |
| chr9_113935917_113936072 | 0.16 |
Ubp1 |
upstream binding protein 1 |
4340 |
0.24 |
| chr9_120541681_120542670 | 0.16 |
Entpd3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
2357 |
0.19 |
| chr5_108627720_108627895 | 0.16 |
Gak |
cyclin G associated kinase |
1616 |
0.22 |
| chr5_113162472_113162792 | 0.16 |
2900026A02Rik |
RIKEN cDNA 2900026A02 gene |
719 |
0.53 |
| chr17_35236085_35236459 | 0.16 |
Atp6v1g2 |
ATPase, H+ transporting, lysosomal V1 subunit G2 |
290 |
0.52 |
| chr5_137293236_137293846 | 0.15 |
Mir8116 |
microRNA 8116 |
239 |
0.76 |
| chr3_67553510_67553744 | 0.15 |
Gm35299 |
predicted gene, 35299 |
202 |
0.91 |
| chr11_61143898_61144049 | 0.15 |
Tnfrsf13b |
tumor necrosis factor receptor superfamily, member 13b |
1308 |
0.36 |
| chr4_43558006_43559275 | 0.15 |
Tln1 |
talin 1 |
180 |
0.86 |
| chr13_73475681_73476013 | 0.15 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
4096 |
0.26 |
| chr5_137644960_137646061 | 0.15 |
Irs3 |
insulin receptor substrate 3 |
27 |
0.93 |
| chr12_12809914_12811298 | 0.15 |
Platr19 |
pluripotency associated transcript 19 |
22621 |
0.16 |
| chr6_67035606_67035757 | 0.15 |
E230016M11Rik |
RIKEN cDNA E230016M11 gene |
918 |
0.38 |
| chr2_137163627_137163778 | 0.15 |
Gm28214 |
predicted gene 28214 |
35281 |
0.19 |
| chr11_120948479_120950230 | 0.15 |
Slc16a3 |
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
273 |
0.84 |
| chr15_58075383_58076476 | 0.15 |
Gm29394 |
predicted gene 29394 |
254 |
0.51 |
| chr9_107887731_107888070 | 0.15 |
Mon1a |
MON1 homolog A, secretory traffciking associated |
251 |
0.83 |
| chr3_34081420_34081639 | 0.14 |
Dnajc19 |
DnaJ heat shock protein family (Hsp40) member C19 |
208 |
0.91 |
| chr9_62341355_62342874 | 0.14 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
39 |
0.98 |
| chr13_101719192_101719926 | 0.14 |
Pik3r1 |
phosphoinositide-3-kinase regulatory subunit 1 |
17455 |
0.2 |
| chr11_45805879_45806041 | 0.14 |
F630206G17Rik |
RIKEN cDNA F630206G17 gene |
2127 |
0.25 |
| chr11_51805212_51805363 | 0.14 |
Sar1b |
secretion associated Ras related GTPase 1B |
41593 |
0.1 |
| chr6_149310794_149310945 | 0.14 |
Resf1 |
retroelement silencing factor 1 |
547 |
0.72 |
| chr8_26329826_26330767 | 0.14 |
Gm31784 |
predicted gene, 31784 |
17962 |
0.15 |
| chr13_21808867_21809156 | 0.14 |
Hist1h2bq |
histone cluster 1, H2bq |
1186 |
0.14 |
| chr2_26960566_26960717 | 0.14 |
Rexo4 |
REX4, 3'-5' exonuclease |
375 |
0.71 |
| chr16_89774022_89774482 | 0.14 |
Tiam1 |
T cell lymphoma invasion and metastasis 1 |
21030 |
0.25 |
| chr1_13573788_13574085 | 0.14 |
Tram1 |
translocating chain-associating membrane protein 1 |
5862 |
0.23 |
| chrX_94178531_94178700 | 0.14 |
Eif2s3x |
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
34036 |
0.13 |
| chr2_76069049_76070775 | 0.13 |
Pde11a |
phosphodiesterase 11A |
63869 |
0.11 |
| chr14_34541288_34541439 | 0.13 |
9230112D13Rik |
RIKEN cDNA 9230112D13 gene |
18562 |
0.11 |
| chr12_59096249_59096457 | 0.13 |
Mia2 |
MIA SH3 domain ER export factor 2 |
553 |
0.61 |
| chr6_37440238_37440988 | 0.13 |
Creb3l2 |
cAMP responsive element binding protein 3-like 2 |
1533 |
0.51 |
| chr5_115161343_115161682 | 0.13 |
Mlec |
malectin |
3333 |
0.13 |
| chr1_71102531_71102932 | 0.13 |
Bard1 |
BRCA1 associated RING domain 1 |
415 |
0.89 |
| chr11_88046995_88047496 | 0.13 |
Srsf1 |
serine and arginine-rich splicing factor 1 |
128 |
0.89 |
| chr7_123370221_123370652 | 0.13 |
Arhgap17 |
Rho GTPase activating protein 17 |
521 |
0.52 |
| chr5_150466366_150466940 | 0.13 |
Fry |
FRY microtubule binding protein |
6007 |
0.12 |
| chr1_135100584_135100735 | 0.13 |
Lgr6 |
leucine-rich repeat-containing G protein-coupled receptor 6 |
4617 |
0.13 |
| chr13_21834970_21835121 | 0.13 |
Hist1h2br |
histone cluster 1 H2br |
1287 |
0.14 |
| chr1_163403739_163404178 | 0.13 |
Gorab |
golgin, RAB6-interacting |
289 |
0.88 |
| chr6_15721115_15721384 | 0.13 |
Mdfic |
MyoD family inhibitor domain containing |
162 |
0.97 |
| chr17_35234891_35235777 | 0.13 |
Atp6v1g2 |
ATPase, H+ transporting, lysosomal V1 subunit G2 |
246 |
0.57 |
| chr7_19149532_19150194 | 0.12 |
Snrpd2 |
small nuclear ribonucleoprotein D2 |
4 |
0.89 |
| chr1_189686405_189686680 | 0.12 |
Gm37239 |
predicted gene, 37239 |
1214 |
0.37 |
| chr11_95307762_95307991 | 0.12 |
Kat7 |
K(lysine) acetyltransferase 7 |
1637 |
0.25 |
| chr13_112196563_112197178 | 0.12 |
Gm37427 |
predicted gene, 37427 |
22323 |
0.16 |
| chr16_55893274_55893677 | 0.12 |
Nxpe3 |
neurexophilin and PC-esterase domain family, member 3 |
1810 |
0.26 |
| chr18_65573679_65573830 | 0.12 |
Zfp532 |
zinc finger protein 532 |
6158 |
0.12 |
| chr1_172500186_172501427 | 0.12 |
Tagln2 |
transgelin 2 |
446 |
0.68 |
| chr4_43000753_43000935 | 0.12 |
Vcp |
valosin containing protein |
337 |
0.78 |
| chr17_71598006_71598722 | 0.12 |
Trmt61b |
tRNA methyltransferase 61B |
111 |
0.93 |
| chr3_146047158_146048273 | 0.12 |
Wdr63 |
WD repeat domain 63 |
530 |
0.75 |
| chr2_93662569_93662820 | 0.12 |
Alx4 |
aristaless-like homeobox 4 |
20306 |
0.21 |
| chr11_78114987_78115399 | 0.12 |
Fam222b |
family with sequence similarity 222, member B |
413 |
0.67 |
| chr4_46043005_46043156 | 0.12 |
Tmod1 |
tropomodulin 1 |
3871 |
0.23 |
| chr18_84858127_84859558 | 0.12 |
Gm16146 |
predicted gene 16146 |
705 |
0.62 |
| chr13_17694404_17694787 | 0.12 |
Sugct |
succinyl-CoA glutarate-CoA transferase |
141 |
0.85 |
| chr8_71394310_71395972 | 0.12 |
Ushbp1 |
USH1 protein network component harmonin binding protein 1 |
644 |
0.48 |
| chr5_21735334_21735486 | 0.12 |
Pmpcb |
peptidase (mitochondrial processing) beta |
1731 |
0.26 |
| chr4_141259880_141260037 | 0.11 |
Gm13055 |
predicted gene 13055 |
6058 |
0.11 |
| chr1_131276506_131276860 | 0.11 |
Ikbke |
inhibitor of kappaB kinase epsilon |
30 |
0.96 |
| chr16_78540388_78540947 | 0.11 |
D16Ertd472e |
DNA segment, Chr 16, ERATO Doi 472, expressed |
19577 |
0.16 |
| chr7_102065578_102066088 | 0.11 |
Xntrpc |
Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
55 |
0.47 |
| chr16_17125146_17125542 | 0.11 |
2610318N02Rik |
RIKEN cDNA 2610318N02 gene |
177 |
0.79 |
| chr15_5243299_5243450 | 0.11 |
Ptger4 |
prostaglandin E receptor 4 (subtype EP4) |
664 |
0.64 |
| chr8_70673716_70674178 | 0.11 |
Lsm4 |
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
515 |
0.58 |
| chr11_48801825_48801976 | 0.11 |
Snord96a |
small nucleolar RNA, C/D box 96A |
132 |
0.85 |
| chr9_35115992_35116896 | 0.11 |
4930581F22Rik |
RIKEN cDNA 4930581F22 gene |
284 |
0.61 |
| chr5_115155040_115155560 | 0.11 |
Mlec |
malectin |
2879 |
0.14 |
| chr1_34439928_34440462 | 0.11 |
Imp4 |
IMP4, U3 small nucleolar ribonucleoprotein |
154 |
0.78 |
| chr11_120241958_120242232 | 0.11 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
5396 |
0.11 |
| chr17_70987811_70988344 | 0.11 |
Myl12b |
myosin, light chain 12B, regulatory |
2453 |
0.16 |
| chr12_117657998_117660727 | 0.11 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
1328 |
0.51 |
| chr17_87283765_87283916 | 0.11 |
Ttc7 |
tetratricopeptide repeat domain 7 |
889 |
0.39 |
| chr4_109678590_109678909 | 0.11 |
Faf1 |
Fas-associated factor 1 |
1752 |
0.31 |
| chr13_9494460_9495468 | 0.11 |
Gm48871 |
predicted gene, 48871 |
49064 |
0.12 |
| chr19_42751879_42752099 | 0.11 |
Pyroxd2 |
pyridine nucleotide-disulphide oxidoreductase domain 2 |
483 |
0.75 |
| chrX_19166017_19167833 | 0.11 |
Gm14636 |
predicted gene 14636 |
307 |
0.9 |
| chr3_121903244_121903507 | 0.11 |
Gm42593 |
predicted gene 42593 |
40533 |
0.12 |
| chr11_115832199_115833263 | 0.10 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
763 |
0.46 |
| chr5_52189299_52190481 | 0.10 |
Dhx15 |
DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
487 |
0.56 |
| chr2_164431743_164432359 | 0.10 |
Sdc4 |
syndecan 4 |
11135 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 0.5 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.2 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 | 0.3 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.1 | 0.2 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.8 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.0 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:1904023 | regulation of fermentation(GO:0043465) regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.0 | 0.0 | GO:0010255 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0043910 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |