Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
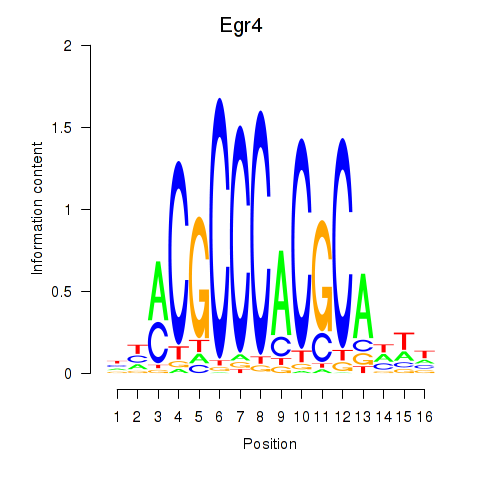
Results for Egr4
Z-value: 0.56
Transcription factors associated with Egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr4
|
ENSMUSG00000071341.3 | early growth response 4 |
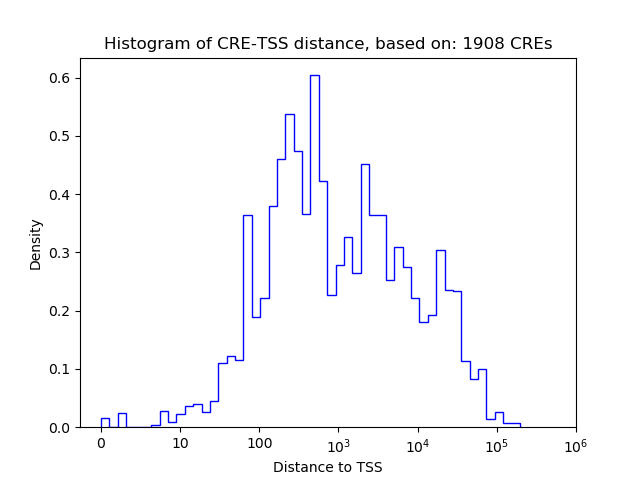
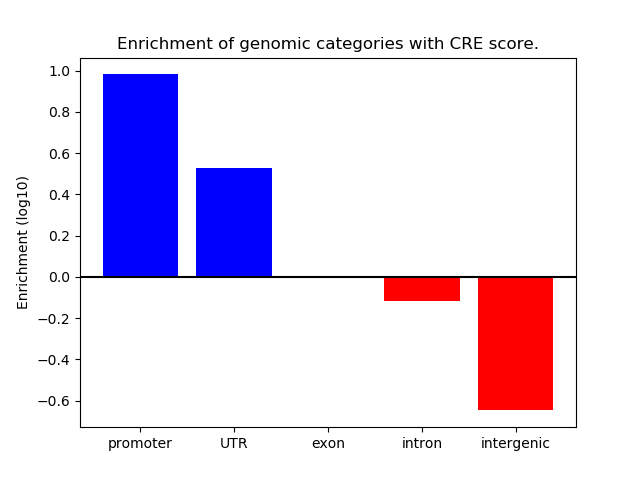
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_85511342_85512700 | Egr4 | 1568 | 0.283108 | -0.52 | 2.1e-05 | Click! |
| chr6_85512870_85513993 | Egr4 | 158 | 0.931004 | -0.50 | 4.8e-05 | Click! |
| chr6_85510634_85510831 | Egr4 | 2857 | 0.177812 | -0.29 | 2.6e-02 | Click! |
| chr6_85518331_85518508 | Egr4 | 4830 | 0.144854 | -0.18 | 1.7e-01 | Click! |
| chr6_85510389_85510540 | Egr4 | 3125 | 0.168991 | 0.01 | 9.5e-01 | Click! |
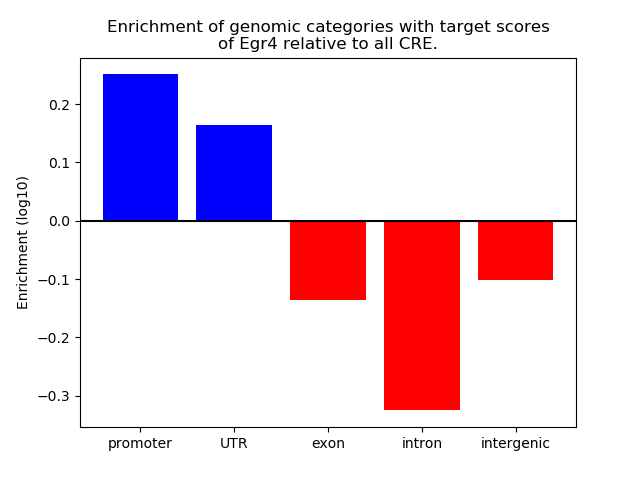
Activity of the Egr4 motif across conditions
Conditions sorted by the z-value of the Egr4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
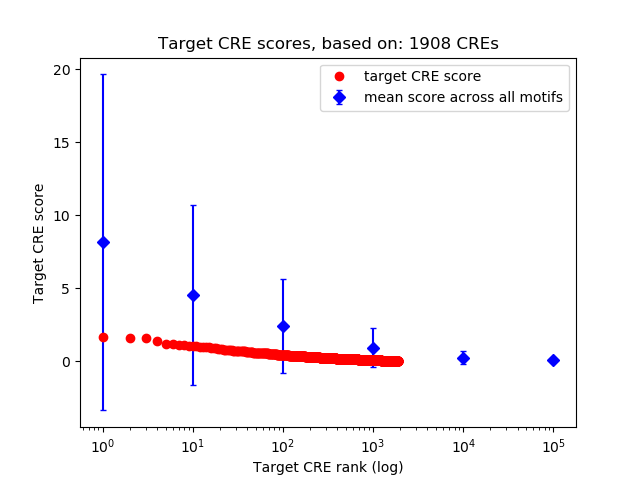
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_142658126_142659753 | 1.69 |
Igf2 |
insulin-like growth factor 2 |
550 |
0.47 |
| chr2_84936571_84938205 | 1.59 |
Slc43a3 |
solute carrier family 43, member 3 |
498 |
0.71 |
| chr17_27825976_27826687 | 1.57 |
Ilrun |
inflammation and lipid regulator with UBA-like and NBR1-like domains |
5683 |
0.13 |
| chr7_142657905_142658056 | 1.37 |
Igf2 |
insulin-like growth factor 2 |
484 |
0.63 |
| chr17_26840808_26841709 | 1.22 |
Nkx2-5 |
NK2 homeobox 5 |
307 |
0.83 |
| chr17_45599296_45600146 | 1.16 |
Slc29a1 |
solute carrier family 29 (nucleoside transporters), member 1 |
115 |
0.92 |
| chr14_101886945_101887764 | 1.10 |
Lmo7 |
LIM domain only 7 |
3235 |
0.35 |
| chr14_63244119_63245953 | 1.08 |
Gata4 |
GATA binding protein 4 |
212 |
0.93 |
| chr17_56123820_56124985 | 1.05 |
Lrg1 |
leucine-rich alpha-2-glycoprotein 1 |
2401 |
0.13 |
| chr8_121088119_121090419 | 1.02 |
Gm27530 |
predicted gene, 27530 |
4563 |
0.13 |
| chr10_60346425_60348008 | 1.01 |
Vsir |
V-set immunoregulatory receptor |
204 |
0.94 |
| chr16_92358680_92359210 | 1.01 |
Kcne1 |
potassium voltage-gated channel, Isk-related subfamily, member 1 |
71 |
0.96 |
| chr12_3806578_3808221 | 1.00 |
Dnmt3a |
DNA methyltransferase 3A |
239 |
0.92 |
| chr1_140180846_140181083 | 0.98 |
Cfh |
complement component factor h |
2316 |
0.37 |
| chr14_69994534_69995596 | 0.95 |
Gm33524 |
predicted gene, 33524 |
62302 |
0.09 |
| chr8_69887012_69889642 | 0.92 |
Yjefn3 |
YjeF N-terminal domain containing 3 |
142 |
0.84 |
| chr5_32745567_32746421 | 0.89 |
Pisd |
phosphatidylserine decarboxylase |
318 |
0.82 |
| chr7_142576289_142578620 | 0.88 |
H19 |
H19, imprinted maternally expressed transcript |
68 |
0.78 |
| chr10_69924337_69924592 | 0.86 |
Ank3 |
ankyrin 3, epithelial |
1031 |
0.69 |
| chr12_31077047_31077429 | 0.86 |
Fam110c |
family with sequence similarity 110, member C |
3377 |
0.17 |
| chr1_185515159_185516225 | 0.82 |
5033404E19Rik |
RIKEN cDNA 5033404E19 gene |
18304 |
0.17 |
| chr4_119188012_119188665 | 0.81 |
Ermap |
erythroblast membrane-associated protein |
409 |
0.69 |
| chr5_101703476_101703804 | 0.79 |
9430085M18Rik |
RIKEN cDNA 9430085M18 gene |
54 |
0.87 |
| chr8_57331786_57332929 | 0.76 |
5033428I22Rik |
RIKEN cDNA 5033428I22 gene |
78 |
0.95 |
| chr11_106485576_106485727 | 0.75 |
Ern1 |
endoplasmic reticulum (ER) to nucleus signalling 1 |
2145 |
0.25 |
| chr2_167367751_167368519 | 0.74 |
B4galt5 |
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
18952 |
0.17 |
| chr16_94722721_94723264 | 0.74 |
Gm41505 |
predicted gene, 41505 |
1098 |
0.52 |
| chr10_127511678_127514192 | 0.74 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
2032 |
0.18 |
| chr2_119237001_119238240 | 0.73 |
Spint1 |
serine protease inhibitor, Kunitz type 1 |
67 |
0.95 |
| chr5_113972560_113973340 | 0.73 |
Ssh1 |
slingshot protein phosphatase 1 |
6194 |
0.14 |
| chr11_75513526_75514670 | 0.73 |
Scarf1 |
scavenger receptor class F, member 1 |
523 |
0.58 |
| chr17_28349544_28350713 | 0.72 |
Tead3 |
TEA domain family member 3 |
218 |
0.87 |
| chr4_154926533_154926891 | 0.71 |
Tnfrsf14 |
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
1365 |
0.31 |
| chr2_32386034_32386242 | 0.71 |
Lcn2 |
lipocalin 2 |
31 |
0.94 |
| chr3_154334782_154335290 | 0.70 |
AI606473 |
expressed sequence AI606473 |
3039 |
0.26 |
| chr12_32060123_32061616 | 0.69 |
Prkar2b |
protein kinase, cAMP dependent regulatory, type II beta |
280 |
0.91 |
| chr2_181458317_181459442 | 0.68 |
Zbtb46 |
zinc finger and BTB domain containing 46 |
500 |
0.67 |
| chr11_102665115_102666490 | 0.68 |
Meioc |
meiosis specific with coiled-coil domain |
179 |
0.9 |
| chr2_147083670_147085642 | 0.67 |
Nkx2-4 |
NK2 homeobox 4 |
789 |
0.65 |
| chr2_170221879_170223257 | 0.66 |
Gm14270 |
predicted gene 14270 |
62467 |
0.12 |
| chr5_120138122_120139268 | 0.66 |
Gm10390 |
predicted gene 10390 |
566 |
0.77 |
| chr3_97628168_97629310 | 0.66 |
Fmo5 |
flavin containing monooxygenase 5 |
65 |
0.96 |
| chr8_84202337_84202869 | 0.66 |
Gm37352 |
predicted gene, 37352 |
1734 |
0.12 |
| chr3_84189998_84191469 | 0.65 |
Trim2 |
tripartite motif-containing 2 |
210 |
0.94 |
| chr15_89168947_89170050 | 0.65 |
Plxnb2 |
plexin B2 |
1190 |
0.27 |
| chr6_117168329_117169820 | 0.60 |
Cxcl12 |
chemokine (C-X-C motif) ligand 12 |
497 |
0.78 |
| chr13_119961525_119962443 | 0.60 |
Gm20784 |
predicted gene, 20784 |
260 |
0.85 |
| chr6_136449235_136449511 | 0.60 |
Gm25882 |
predicted gene, 25882 |
31561 |
0.1 |
| chr15_103253562_103255772 | 0.60 |
Nfe2 |
nuclear factor, erythroid derived 2 |
605 |
0.57 |
| chr5_83352776_83353067 | 0.59 |
Tecrl |
trans-2,3-enoyl-CoA reductase-like |
2035 |
0.43 |
| chr7_110774911_110775345 | 0.58 |
Ampd3 |
adenosine monophosphate deaminase 3 |
884 |
0.5 |
| chr10_108434849_108435510 | 0.58 |
Gm36283 |
predicted gene, 36283 |
1486 |
0.41 |
| chr14_53729532_53730255 | 0.57 |
Trav13-3 |
T cell receptor alpha variable 13-3 |
335 |
0.84 |
| chr14_53337071_53337774 | 0.57 |
Gm43650 |
predicted gene 43650 |
6500 |
0.19 |
| chr5_137569532_137569798 | 0.57 |
Tfr2 |
transferrin receptor 2 |
175 |
0.85 |
| chr10_79897288_79897960 | 0.56 |
Med16 |
mediator complex subunit 16 |
2061 |
0.1 |
| chr19_46304366_46306224 | 0.56 |
4833438C02Rik |
RIKEN cDNA 4833438C02 gene |
310 |
0.52 |
| chr10_119408311_119408592 | 0.56 |
Gm40778 |
predicted gene, 40778 |
2771 |
0.18 |
| chr18_75494415_75495639 | 0.56 |
Gm10532 |
predicted gene 10532 |
19618 |
0.24 |
| chr2_9882196_9886301 | 0.55 |
9230102O04Rik |
RIKEN cDNA 9230102O04 gene |
255 |
0.84 |
| chr4_133659590_133660887 | 0.55 |
Zdhhc18 |
zinc finger, DHHC domain containing 18 |
10084 |
0.12 |
| chr7_19697995_19699396 | 0.55 |
Apoe |
apolipoprotein E |
74 |
0.92 |
| chr2_162947675_162948537 | 0.55 |
L3mbtl1 |
L3MBTL1 histone methyl-lysine binding protein |
101 |
0.95 |
| chr9_55283384_55284127 | 0.55 |
Nrg4 |
neuregulin 4 |
130 |
0.96 |
| chr3_144380827_144381780 | 0.55 |
Gm5857 |
predicted gene 5857 |
46369 |
0.13 |
| chr16_42180798_42180949 | 0.54 |
Gm49737 |
predicted gene, 49737 |
4044 |
0.24 |
| chr13_43123788_43124308 | 0.54 |
Phactr1 |
phosphatase and actin regulator 1 |
1176 |
0.42 |
| chr12_105981623_105981921 | 0.54 |
Vrk1 |
vaccinia related kinase 1 |
28456 |
0.18 |
| chr2_77401485_77402614 | 0.53 |
Gm13944 |
predicted gene 13944 |
60620 |
0.11 |
| chr2_105125289_105128976 | 0.53 |
Wt1 |
Wilms tumor 1 homolog |
78 |
0.91 |
| chr14_53032863_53033592 | 0.52 |
Gm30214 |
predicted gene, 30214 |
6239 |
0.17 |
| chr15_76666348_76670076 | 0.51 |
Foxh1 |
forkhead box H1 |
1590 |
0.15 |
| chr7_25686795_25687582 | 0.50 |
Tgfb1 |
transforming growth factor, beta 1 |
186 |
0.88 |
| chr3_89251147_89252390 | 0.49 |
Krtcap2 |
keratinocyte associated protein 2 |
2901 |
0.08 |
| chr9_79975154_79975866 | 0.49 |
Filip1 |
filamin A interacting protein 1 |
2149 |
0.3 |
| chr5_75046578_75047197 | 0.49 |
Gm18593 |
predicted gene, 18593 |
1592 |
0.25 |
| chr6_50030123_50030453 | 0.48 |
Gm3455 |
predicted gene 3455 |
20898 |
0.24 |
| chr3_108225902_108227171 | 0.48 |
Sypl2 |
synaptophysin-like 2 |
107 |
0.67 |
| chr15_79893596_79894135 | 0.47 |
Apobec3 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
1158 |
0.33 |
| chr11_70228540_70230880 | 0.47 |
Bcl6b |
B cell CLL/lymphoma 6, member B |
29 |
0.93 |
| chr11_106709558_106710158 | 0.47 |
Pecam1 |
platelet/endothelial cell adhesion molecule 1 |
5088 |
0.16 |
| chr13_104822344_104822827 | 0.47 |
Srek1ip1 |
splicing regulatory glutamine/lysine-rich protein 1interacting protein 1 |
4073 |
0.23 |
| chr17_48449646_48450503 | 0.47 |
Tspo2 |
translocator protein 2 |
1 |
0.96 |
| chr6_83762249_83763628 | 0.47 |
1700124L16Rik |
RIKEN cDNA 1700124L16 gene |
73 |
0.72 |
| chr6_3967558_3968787 | 0.47 |
Tfpi2 |
tissue factor pathway inhibitor 2 |
204 |
0.94 |
| chr13_120100052_120100925 | 0.46 |
Gm21378 |
predicted gene, 21378 |
156 |
0.93 |
| chr13_45513570_45514179 | 0.46 |
Gmpr |
guanosine monophosphate reductase |
34 |
0.98 |
| chr2_148043571_148045987 | 0.46 |
Foxa2 |
forkhead box A2 |
685 |
0.65 |
| chr5_37901474_37902022 | 0.46 |
Gm20052 |
predicted gene, 20052 |
7041 |
0.23 |
| chr1_155039490_155039693 | 0.45 |
Gm29441 |
predicted gene 29441 |
7852 |
0.18 |
| chr5_122774691_122775143 | 0.44 |
Camkk2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
4364 |
0.14 |
| chr16_78249394_78249789 | 0.44 |
Gm49591 |
predicted gene, 49591 |
1462 |
0.35 |
| chr7_128301638_128302100 | 0.44 |
BC017158 |
cDNA sequence BC017158 |
3699 |
0.12 |
| chr7_4520460_4522358 | 0.44 |
Tnni3 |
troponin I, cardiac 3 |
842 |
0.35 |
| chr2_152962069_152963079 | 0.44 |
Ttll9 |
tubulin tyrosine ligase-like family, member 9 |
63 |
0.96 |
| chr11_61683750_61685180 | 0.44 |
Fam83g |
family with sequence similarity 83, member G |
46 |
0.97 |
| chr18_64485275_64485762 | 0.43 |
Fech |
ferrochelatase |
3423 |
0.2 |
| chr8_94984127_94985009 | 0.43 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
98 |
0.95 |
| chr2_68873756_68874637 | 0.43 |
Cers6 |
ceramide synthase 6 |
12610 |
0.14 |
| chr6_54698722_54699891 | 0.43 |
Gm44008 |
predicted gene, 44008 |
5660 |
0.18 |
| chr11_96282910_96285155 | 0.42 |
Hoxb8 |
homeobox B8 |
1307 |
0.19 |
| chr1_156938608_156939649 | 0.42 |
Ralgps2 |
Ral GEF with PH domain and SH3 binding motif 2 |
6 |
0.94 |
| chr9_116172528_116172709 | 0.42 |
Tgfbr2 |
transforming growth factor, beta receptor II |
2647 |
0.26 |
| chr17_57264956_57265836 | 0.42 |
Gm46575 |
predicted gene, 46575 |
2526 |
0.17 |
| chr15_28024567_28025840 | 0.42 |
Trio |
triple functional domain (PTPRF interacting) |
249 |
0.93 |
| chr4_154635108_154637998 | 0.42 |
Prdm16 |
PR domain containing 16 |
244 |
0.83 |
| chr11_49087855_49088820 | 0.42 |
Ifi47 |
interferon gamma inducible protein 47 |
73 |
0.66 |
| chr8_84703616_84705950 | 0.41 |
Nfix |
nuclear factor I/X |
2933 |
0.13 |
| chr4_48407474_48407947 | 0.41 |
Invs |
inversin |
25522 |
0.19 |
| chr5_35391428_35392811 | 0.41 |
4930442P19Rik |
RIKEN cDNA 4930442P19 gene |
207 |
0.9 |
| chr2_156840101_156841614 | 0.41 |
Tgif2 |
TGFB-induced factor homeobox 2 |
165 |
0.85 |
| chr4_152158170_152159968 | 0.41 |
Hes2 |
hes family bHLH transcription factor 2 |
202 |
0.88 |
| chr11_83065104_83067047 | 0.41 |
Slfn2 |
schlafen 2 |
963 |
0.31 |
| chr7_112742800_112743435 | 0.40 |
Tead1 |
TEA domain family member 1 |
1072 |
0.55 |
| chr3_90450566_90451614 | 0.40 |
Gm16048 |
predicted gene 16048 |
493 |
0.47 |
| chr4_133649781_133650054 | 0.40 |
Zdhhc18 |
zinc finger, DHHC domain containing 18 |
237 |
0.88 |
| chr7_143100274_143100425 | 0.40 |
Trpm5 |
transient receptor potential cation channel, subfamily M, member 5 |
5707 |
0.11 |
| chr19_56115631_56116270 | 0.39 |
Gm31912 |
predicted gene, 31912 |
9640 |
0.26 |
| chr9_64044996_64045309 | 0.39 |
Gm25606 |
predicted gene, 25606 |
3344 |
0.16 |
| chr16_22920044_22920195 | 0.39 |
Fetub |
fetuin beta |
118 |
0.94 |
| chr17_23674874_23675429 | 0.39 |
Thoc6 |
THO complex 6 |
1269 |
0.15 |
| chr14_100284227_100286288 | 0.39 |
Klf12 |
Kruppel-like factor 12 |
578 |
0.59 |
| chr7_35119262_35120228 | 0.39 |
Gm45091 |
predicted gene 45091 |
140 |
0.78 |
| chr17_48300015_48301474 | 0.39 |
Treml2 |
triggering receptor expressed on myeloid cells-like 2 |
358 |
0.8 |
| chr17_47436222_47437660 | 0.39 |
AI661453 |
expressed sequence AI661453 |
136 |
0.79 |
| chr1_167269338_167269683 | 0.38 |
Uck2 |
uridine-cytidine kinase 2 |
15091 |
0.11 |
| chr1_134513098_134514080 | 0.38 |
Gm15454 |
predicted gene 15454 |
4492 |
0.12 |
| chr8_23038726_23039184 | 0.38 |
Ank1 |
ankyrin 1, erythroid |
3724 |
0.22 |
| chr16_92696683_92697461 | 0.38 |
Runx1 |
runt related transcription factor 1 |
253 |
0.95 |
| chr17_71332316_71332674 | 0.38 |
Gm4566 |
predicted gene 4566 |
9271 |
0.12 |
| chr8_71665120_71666309 | 0.38 |
Unc13a |
unc-13 homolog A |
2368 |
0.14 |
| chr10_43667858_43668095 | 0.38 |
Gm34481 |
predicted gene, 34481 |
8658 |
0.13 |
| chr2_4298620_4299406 | 0.38 |
Frmd4a |
FERM domain containing 4A |
1505 |
0.32 |
| chr5_142938090_142938561 | 0.38 |
Fscn1 |
fascin actin-bundling protein 1 |
22018 |
0.13 |
| chr15_57040178_57040584 | 0.38 |
Gm2582 |
predicted gene 2582 |
33757 |
0.23 |
| chr2_156780383_156781526 | 0.37 |
Myl9 |
myosin, light polypeptide 9, regulatory |
2437 |
0.18 |
| chr5_101665386_101665820 | 0.37 |
Nkx6-1 |
NK6 homeobox 1 |
607 |
0.74 |
| chr4_133055332_133056821 | 0.37 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
10173 |
0.19 |
| chr19_32866766_32867884 | 0.37 |
Pten |
phosphatase and tensin homolog |
56100 |
0.14 |
| chr3_131110318_131112630 | 0.37 |
Lef1os1 |
LEF1 opposite strand RNA 1 |
616 |
0.56 |
| chr9_108081761_108082036 | 0.37 |
Mir7088 |
microRNA 7088 |
400 |
0.57 |
| chr12_57542139_57543972 | 0.37 |
Foxa1 |
forkhead box A1 |
3066 |
0.2 |
| chr5_116446483_116447070 | 0.37 |
Srrm4 |
serine/arginine repetitive matrix 4 |
68 |
0.96 |
| chr9_66157901_66159171 | 0.37 |
Dapk2 |
death-associated protein kinase 2 |
301 |
0.9 |
| chr6_39724228_39725649 | 0.37 |
Braf |
Braf transforming gene |
255 |
0.93 |
| chr10_70119512_70120499 | 0.37 |
Ccdc6 |
coiled-coil domain containing 6 |
22884 |
0.22 |
| chr2_174298225_174300890 | 0.36 |
Gnas |
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
1698 |
0.27 |
| chr4_123233207_123234595 | 0.36 |
Heyl |
hairy/enhancer-of-split related with YRPW motif-like |
48 |
0.96 |
| chr10_18055567_18056708 | 0.36 |
Reps1 |
RalBP1 associated Eps domain containing protein |
133 |
0.96 |
| chr1_191855077_191855680 | 0.36 |
Nek2 |
NIMA (never in mitosis gene a)-related expressed kinase 2 |
28230 |
0.12 |
| chr2_25195300_25197035 | 0.36 |
Tor4a |
torsin family 4, member A |
592 |
0.43 |
| chr14_114857722_114857913 | 0.36 |
Gm49010 |
predicted gene, 49010 |
18693 |
0.16 |
| chr11_22376584_22377158 | 0.36 |
Rpsa-ps3 |
ribosomal protein SA, pseudogene 3 |
9421 |
0.22 |
| chr1_74035856_74036007 | 0.36 |
Tns1 |
tensin 1 |
1202 |
0.53 |
| chr14_76532606_76533942 | 0.36 |
E130202H07Rik |
RIKEN cDNA E130202H07 gene |
6658 |
0.2 |
| chr4_98923707_98924979 | 0.36 |
Usp1 |
ubiquitin specific peptidase 1 |
379 |
0.86 |
| chr2_36230119_36231737 | 0.35 |
Ptgs1 |
prostaglandin-endoperoxide synthase 1 |
372 |
0.79 |
| chr11_19827943_19828094 | 0.35 |
Gm12029 |
predicted gene 12029 |
44366 |
0.15 |
| chr5_5261065_5262223 | 0.35 |
Cdk14 |
cyclin-dependent kinase 14 |
3665 |
0.25 |
| chr11_95337114_95338152 | 0.35 |
Gm11521 |
predicted gene 11521 |
225 |
0.77 |
| chr3_36531782_36532903 | 0.35 |
Gm11549 |
predicted gene 11549 |
11 |
0.96 |
| chr15_80061908_80062059 | 0.35 |
Snord83b |
small nucleolar RNA, C/D box 83B |
16608 |
0.09 |
| chr3_152477886_152478987 | 0.35 |
Ak5 |
adenylate kinase 5 |
79 |
0.97 |
| chr7_16844794_16846016 | 0.35 |
Prkd2 |
protein kinase D2 |
172 |
0.9 |
| chr7_15948288_15949198 | 0.34 |
Nop53 |
NOP53 ribosome biogenesis factor |
2669 |
0.14 |
| chr18_50029220_50029371 | 0.34 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
1723 |
0.39 |
| chr11_114851248_114852672 | 0.34 |
Gprc5c |
G protein-coupled receptor, family C, group 5, member C |
80 |
0.96 |
| chr5_23417846_23419038 | 0.34 |
5031425E22Rik |
RIKEN cDNA 5031425E22 gene |
14867 |
0.12 |
| chr7_19769512_19770898 | 0.34 |
Bcam |
basal cell adhesion molecule |
290 |
0.54 |
| chr2_124609830_124611153 | 0.34 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
82 |
0.99 |
| chr9_37421331_37421587 | 0.33 |
Robo3 |
roundabout guidance receptor 3 |
1144 |
0.37 |
| chr2_153528339_153529939 | 0.33 |
Nol4l |
nucleolar protein 4-like |
832 |
0.63 |
| chr9_79714238_79715732 | 0.33 |
Col12a1 |
collagen, type XII, alpha 1 |
3509 |
0.22 |
| chr9_50751156_50752468 | 0.33 |
Cryab |
crystallin, alpha B |
65 |
0.86 |
| chr5_52254526_52255173 | 0.33 |
Gm43180 |
predicted gene 43180 |
15513 |
0.15 |
| chr1_174171570_174172092 | 0.33 |
Spta1 |
spectrin alpha, erythrocytic 1 |
945 |
0.37 |
| chr11_4031787_4032434 | 0.33 |
Sec14l4 |
SEC14-like lipid binding 4 |
268 |
0.85 |
| chr13_12680135_12680751 | 0.33 |
Gm48455 |
predicted gene, 48455 |
503 |
0.38 |
| chr6_52174920_52176658 | 0.33 |
Hoxaas3 |
Hoxa cluster antisense RNA 3 |
554 |
0.41 |
| chr17_35978858_35980224 | 0.33 |
Prr3 |
proline-rich polypeptide 3 |
15 |
0.78 |
| chr2_119618746_119619194 | 0.33 |
Nusap1 |
nucleolar and spindle associated protein 1 |
241 |
0.69 |
| chr11_94455571_94456396 | 0.33 |
Cacna1g |
calcium channel, voltage-dependent, T type, alpha 1G subunit |
17658 |
0.13 |
| chr5_115119516_115119726 | 0.33 |
Acads |
acyl-Coenzyme A dehydrogenase, short chain |
275 |
0.82 |
| chr10_75407240_75407611 | 0.33 |
Upb1 |
ureidopropionase, beta |
467 |
0.8 |
| chr2_132102631_132103055 | 0.32 |
Gm14052 |
predicted gene 14052 |
2974 |
0.23 |
| chr2_25577192_25580600 | 0.32 |
Ajm1 |
apical junction component 1 |
1001 |
0.25 |
| chr19_46959766_46960228 | 0.32 |
Nt5c2 |
5'-nucleotidase, cytosolic II |
1923 |
0.31 |
| chrX_36025305_36025924 | 0.32 |
Gm50468 |
predicted gene, 50468 |
434 |
0.85 |
| chrX_77432774_77434116 | 0.32 |
Gm5937 |
predicted gene 5937 |
23141 |
0.22 |
| chr2_126498850_126499812 | 0.32 |
Atp8b4 |
ATPase, class I, type 8B, member 4 |
516 |
0.83 |
| chr5_116422576_116423267 | 0.31 |
Hspb8 |
heat shock protein 8 |
57 |
0.96 |
| chr2_75776376_75777638 | 0.31 |
Gm13657 |
predicted gene 13657 |
181 |
0.93 |
| chr7_89903398_89904159 | 0.31 |
Ccdc81 |
coiled-coil domain containing 81 |
149 |
0.95 |
| chrX_75232876_75233354 | 0.31 |
Gm6039 |
predicted gene 6039 |
52 |
0.97 |
| chr2_37786020_37786930 | 0.31 |
Crb2 |
crumbs family member 2 |
4696 |
0.25 |
| chr18_84087366_84088497 | 0.31 |
Zadh2 |
zinc binding alcohol dehydrogenase, domain containing 2 |
132 |
0.63 |
| chr12_69582084_69583353 | 0.31 |
Vcpkmt |
valosin containing protein lysine (K) methyltransferase |
217 |
0.9 |
| chr8_117256613_117257991 | 0.31 |
Cmip |
c-Maf inducing protein |
185 |
0.96 |
| chr5_125521168_125521465 | 0.31 |
Aacs |
acetoacetyl-CoA synthetase |
6073 |
0.17 |
| chr7_45233983_45235751 | 0.31 |
Cd37 |
CD37 antigen |
591 |
0.44 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 1.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.3 | 1.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 1.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.9 | GO:1900212 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.2 | 0.9 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.2 | 0.6 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.2 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 0.3 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.2 | 0.5 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 1.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 0.8 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.4 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.5 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.7 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.3 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.7 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 1.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.1 | 0.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.6 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.6 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.3 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.2 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.0 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.0 | 0.1 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.2 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.3 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.1 | GO:0071655 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0072162 | metanephric mesenchymal cell differentiation(GO:0072162) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0071107 | response to parathyroid hormone(GO:0071107) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0015781 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.3 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0046040 | IMP metabolic process(GO:0046040) |
| 0.0 | 0.3 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.0 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 | 0.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0046719 | regulation by virus of viral protein levels in host cell(GO:0046719) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.0 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.0 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.0 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.0 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 2.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.2 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 2.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0034838 | enoyl-[acyl-carrier-protein] reductase activity(GO:0016631) 2,3-dihydroxy-2,3-dihydro-phenylpropionate dehydrogenase activity(GO:0018498) cis-2,3-dihydrodiol DDT dehydrogenase activity(GO:0018499) trans-9R,10R-dihydrodiolphenanthrene dehydrogenase activity(GO:0018500) cis-chlorobenzene dihydrodiol dehydrogenase activity(GO:0018501) 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase activity(GO:0018502) trans-1,2-dihydrodiolphenanthrene dehydrogenase activity(GO:0018503) 3,4-dihydroxy-3,4-dihydrofluorene dehydrogenase activity(GO:0034790) benzo(a)pyrene-trans-11,12-dihydrodiol dehydrogenase activity(GO:0034805) benzo(a)pyrene-cis-4,5-dihydrodiol dehydrogenase activity(GO:0034809) citronellyl-CoA dehydrogenase activity(GO:0034824) menthone dehydrogenase activity(GO:0034838) phthalate 3,4-cis-dihydrodiol dehydrogenase activity(GO:0034912) cinnamate reductase activity(GO:0043786) NADPH-dependent curcumin reductase activity(GO:0052849) NADPH-dependent dihydrocurcumin reductase activity(GO:0052850) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0034547 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.4 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0015254 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0071813 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0043738 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |