Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
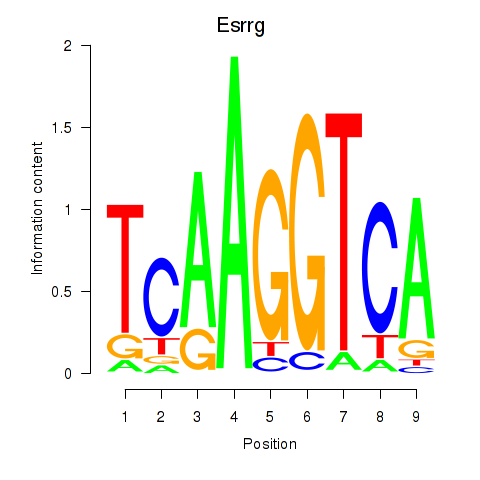
Results for Esrrg
Z-value: 0.54
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSMUSG00000026610.7 | estrogen-related receptor gamma |
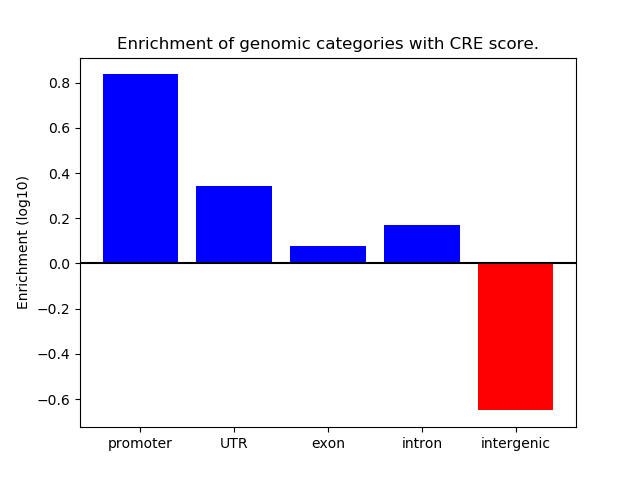
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_187997341_187998650 | Esrrg | 127 | 0.972695 | 0.49 | 6.3e-05 | Click! |
| chr1_187604626_187606001 | Esrrg | 3478 | 0.282480 | 0.45 | 3.3e-04 | Click! |
| chr1_188001052_188001305 | Esrrg | 3310 | 0.295637 | 0.44 | 4.3e-04 | Click! |
| chr1_187999916_188000681 | Esrrg | 2430 | 0.345336 | 0.42 | 8.9e-04 | Click! |
| chr1_187998988_187999336 | Esrrg | 1294 | 0.524252 | 0.42 | 9.6e-04 | Click! |
Activity of the Esrrg motif across conditions
Conditions sorted by the z-value of the Esrrg motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
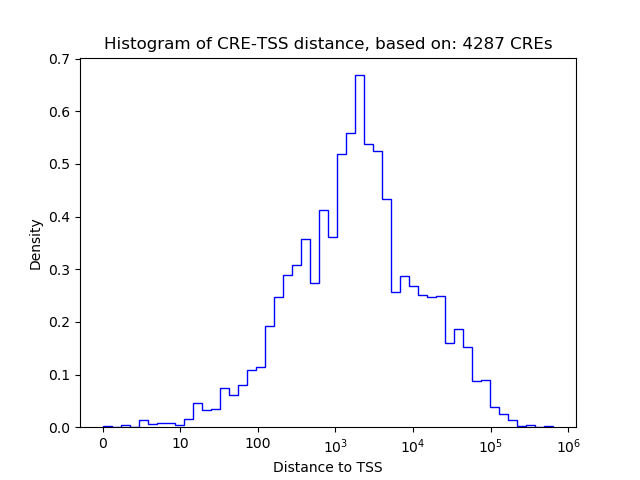
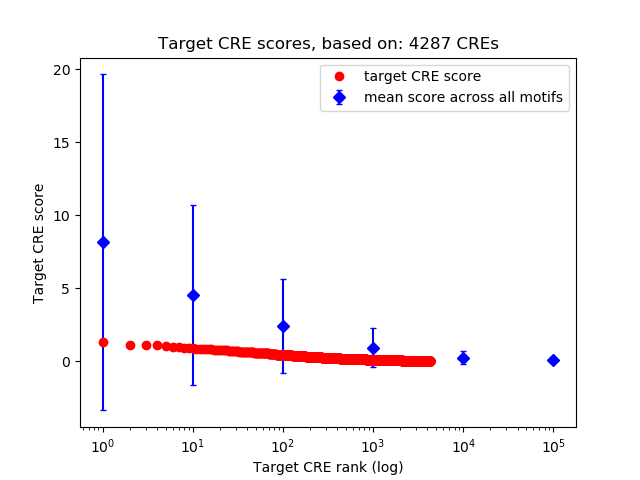
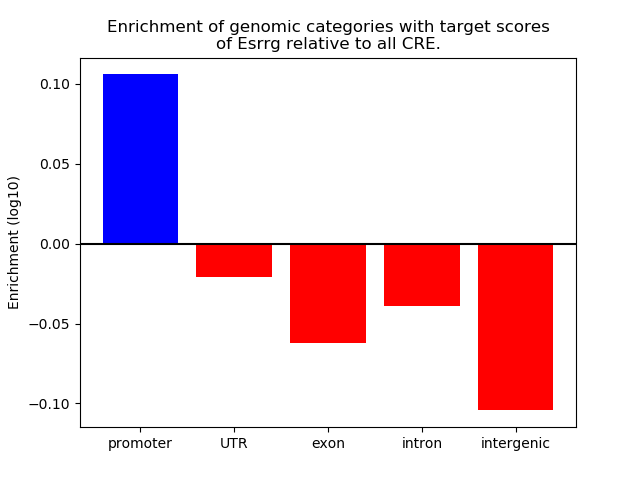
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_99513882_99514334 | 1.35 |
Map1b |
microtubule-associated protein 1B |
2410 |
0.23 |
| chr2_181715341_181715994 | 1.12 |
Oprl1 |
opioid receptor-like 1 |
35 |
0.95 |
| chr13_83722679_83723219 | 1.09 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
1568 |
0.29 |
| chr2_91119045_91119896 | 1.09 |
Mybpc3 |
myosin binding protein C, cardiac |
1326 |
0.3 |
| chr10_3368375_3368998 | 1.08 |
Ppp1r14c |
protein phosphatase 1, regulatory inhibitor subunit 14C |
2142 |
0.36 |
| chr14_20663037_20663711 | 0.99 |
Synpo2l |
synaptopodin 2-like |
1200 |
0.26 |
| chr10_69536775_69537233 | 0.97 |
Ank3 |
ankyrin 3, epithelial |
2782 |
0.32 |
| chr12_86680036_86680970 | 0.93 |
Vash1 |
vasohibin 1 |
1803 |
0.28 |
| chr5_135248496_135249658 | 0.89 |
Fzd9 |
frizzled class receptor 9 |
2153 |
0.21 |
| chr9_110765100_110765787 | 0.88 |
Myl3 |
myosin, light polypeptide 3 |
418 |
0.74 |
| chr10_30840531_30840931 | 0.86 |
Hey2 |
hairy/enhancer-of-split related with YRPW motif 2 |
1284 |
0.43 |
| chr7_126274286_126274741 | 0.85 |
Sbk1 |
SH3-binding kinase 1 |
1113 |
0.38 |
| chr18_24711119_24711270 | 0.84 |
Fhod3 |
formin homology 2 domain containing 3 |
1749 |
0.28 |
| chr9_56535349_56535646 | 0.84 |
Gm47176 |
predicted gene, 47176 |
4408 |
0.18 |
| chr1_59766668_59766947 | 0.83 |
Bmpr2 |
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
3407 |
0.19 |
| chr9_56864653_56866648 | 0.82 |
Cspg4 |
chondroitin sulfate proteoglycan 4 |
617 |
0.51 |
| chr4_143300007_143300403 | 0.81 |
Pdpn |
podoplanin |
641 |
0.68 |
| chr9_97017174_97017806 | 0.81 |
Gm16010 |
predicted gene 16010 |
16 |
0.96 |
| chr17_72921491_72924008 | 0.80 |
Lbh |
limb-bud and heart |
1561 |
0.47 |
| chr6_85371815_85372854 | 0.79 |
Rab11fip5 |
RAB11 family interacting protein 5 (class I) |
2230 |
0.24 |
| chr10_80180425_80180593 | 0.77 |
Efna2 |
ephrin A2 |
1027 |
0.3 |
| chr14_34822420_34823516 | 0.77 |
Grid1 |
glutamate receptor, ionotropic, delta 1 |
2860 |
0.27 |
| chr14_101886945_101887764 | 0.76 |
Lmo7 |
LIM domain only 7 |
3235 |
0.35 |
| chr13_83737592_83739114 | 0.74 |
Gm33366 |
predicted gene, 33366 |
182 |
0.66 |
| chr16_94722721_94723264 | 0.73 |
Gm41505 |
predicted gene, 41505 |
1098 |
0.52 |
| chr8_95703051_95704038 | 0.72 |
Ndrg4 |
N-myc downstream regulated gene 4 |
474 |
0.68 |
| chr10_13322058_13322496 | 0.72 |
Phactr2 |
phosphatase and actin regulator 2 |
2012 |
0.42 |
| chr10_111901253_111901501 | 0.71 |
Gm47880 |
predicted gene, 47880 |
12363 |
0.16 |
| chr9_30920196_30920963 | 0.70 |
Adamts15 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 15 |
371 |
0.88 |
| chr18_60646141_60646566 | 0.68 |
Synpo |
synaptopodin |
1919 |
0.32 |
| chr7_141428600_141429722 | 0.68 |
Cend1 |
cell cycle exit and neuronal differentiation 1 |
190 |
0.82 |
| chrX_134295383_134296942 | 0.67 |
Tmem35a |
transmembrane protein 35A |
937 |
0.52 |
| chr2_170731776_170732705 | 0.66 |
Dok5 |
docking protein 5 |
433 |
0.88 |
| chr2_114048099_114048464 | 0.65 |
Actc1 |
actin, alpha, cardiac muscle 1 |
4606 |
0.18 |
| chr6_38252574_38252916 | 0.65 |
D630045J12Rik |
RIKEN cDNA D630045J12 gene |
1210 |
0.46 |
| chr17_81737002_81738450 | 0.64 |
Slc8a1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
651 |
0.81 |
| chr5_114147552_114147966 | 0.64 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
1224 |
0.34 |
| chr10_127261534_127261755 | 0.64 |
Kif5a |
kinesin family member 5A |
1436 |
0.2 |
| chr1_169746128_169746823 | 0.63 |
Rgs4 |
regulator of G-protein signaling 4 |
1148 |
0.55 |
| chr14_55056074_55056891 | 0.63 |
Gm20687 |
predicted gene 20687 |
989 |
0.3 |
| chr9_66512511_66514532 | 0.63 |
Fbxl22 |
F-box and leucine-rich repeat protein 22 |
1088 |
0.47 |
| chr2_91126778_91126929 | 0.62 |
Mybpc3 |
myosin binding protein C, cardiac |
8709 |
0.11 |
| chr4_43406531_43407612 | 0.62 |
Rusc2 |
RUN and SH3 domain containing 2 |
258 |
0.87 |
| chr8_110722042_110722638 | 0.61 |
Mtss2 |
MTSS I-BAR domain containing 2 |
864 |
0.55 |
| chr15_25942778_25943455 | 0.61 |
Retreg1 |
reticulophagy regulator 1 |
437 |
0.83 |
| chr11_111067503_111068134 | 0.61 |
Kcnj2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
1654 |
0.5 |
| chr11_68390483_68390938 | 0.61 |
Ntn1 |
netrin 1 |
3698 |
0.27 |
| chr4_120813989_120815034 | 0.59 |
Nfyc |
nuclear transcription factor-Y gamma |
1201 |
0.36 |
| chr7_87585368_87585534 | 0.59 |
Grm5 |
glutamate receptor, metabotropic 5 |
1053 |
0.67 |
| chr19_10342009_10342277 | 0.59 |
Dagla |
diacylglycerol lipase, alpha |
37266 |
0.11 |
| chr5_104461026_104461334 | 0.59 |
Pkd2 |
polycystin 2, transient receptor potential cation channel |
1610 |
0.34 |
| chr18_47525528_47525697 | 0.58 |
Gm5095 |
predicted gene 5095 |
12123 |
0.21 |
| chr11_98929351_98930679 | 0.58 |
Rara |
retinoic acid receptor, alpha |
2197 |
0.18 |
| chr9_65678023_65678538 | 0.58 |
Oaz2 |
ornithine decarboxylase antizyme 2 |
1689 |
0.32 |
| chr7_19414891_19415097 | 0.58 |
Ckm |
creatine kinase, muscle |
3931 |
0.08 |
| chrX_162025397_162025772 | 0.57 |
Gm26317 |
predicted gene, 26317 |
43873 |
0.17 |
| chr12_100907442_100908590 | 0.57 |
Gpr68 |
G protein-coupled receptor 68 |
165 |
0.92 |
| chr11_3368719_3370424 | 0.56 |
Limk2 |
LIM motif-containing protein kinase 2 |
409 |
0.76 |
| chr8_12926230_12928559 | 0.56 |
Mcf2l |
mcf.2 transforming sequence-like |
762 |
0.52 |
| chr17_32634501_32635224 | 0.56 |
Cyp4f37 |
cytochrome P450, family 4, subfamily f, polypeptide 37 |
160 |
0.92 |
| chr8_82865753_82865928 | 0.55 |
Rnf150 |
ring finger protein 150 |
2026 |
0.38 |
| chr2_114057084_114058386 | 0.55 |
C130080G10Rik |
RIKEN cDNA C130080G10 gene |
1422 |
0.37 |
| chr6_114041186_114042349 | 0.55 |
Atp2b2 |
ATPase, Ca++ transporting, plasma membrane 2 |
248 |
0.91 |
| chr1_191720952_191721137 | 0.55 |
Lpgat1 |
lysophosphatidylglycerol acyltransferase 1 |
2523 |
0.25 |
| chr9_107708227_107708656 | 0.55 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
1038 |
0.34 |
| chr13_117603755_117604285 | 0.55 |
Hcn1 |
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
1540 |
0.54 |
| chr9_121792003_121792549 | 0.54 |
Hhatl |
hedgehog acyltransferase-like |
231 |
0.84 |
| chr3_17789318_17789657 | 0.53 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
434 |
0.83 |
| chr6_128032492_128033212 | 0.53 |
Tspan9 |
tetraspanin 9 |
1740 |
0.36 |
| chr18_69523224_69524043 | 0.52 |
Tcf4 |
transcription factor 4 |
1183 |
0.58 |
| chr5_69541689_69541957 | 0.52 |
Yipf7 |
Yip1 domain family, member 7 |
766 |
0.59 |
| chr5_138992944_138993679 | 0.51 |
Pdgfa |
platelet derived growth factor, alpha |
970 |
0.48 |
| chr10_9092809_9092960 | 0.50 |
Gm48743 |
predicted gene, 48743 |
21409 |
0.23 |
| chr7_125831269_125831420 | 0.50 |
D430042O09Rik |
RIKEN cDNA D430042O09 gene |
1677 |
0.48 |
| chr10_80151643_80151976 | 0.49 |
Midn |
midnolin |
571 |
0.53 |
| chr1_77080723_77080989 | 0.49 |
Gm816 |
predicted gene 816 |
53064 |
0.14 |
| chr15_76519928_76521866 | 0.49 |
Scrt1 |
scratch family zinc finger 1 |
1005 |
0.28 |
| chr18_33460259_33461386 | 0.49 |
Nrep |
neuronal regeneration related protein |
2613 |
0.29 |
| chr14_61257260_61257428 | 0.49 |
Sgcg |
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
1080 |
0.45 |
| chr12_86823362_86823513 | 0.47 |
Gm10095 |
predicted gene 10095 |
23030 |
0.17 |
| chr3_107516925_107518261 | 0.47 |
Slc6a17 |
solute carrier family 6 (neurotransmitter transporter), member 17 |
156 |
0.95 |
| chr14_105333345_105334016 | 0.47 |
Ndfip2 |
Nedd4 family interacting protein 2 |
39134 |
0.13 |
| chr6_120483288_120483595 | 0.46 |
Il17ra |
interleukin 17 receptor A |
9838 |
0.13 |
| chrX_157704254_157705325 | 0.46 |
Smpx |
small muscle protein, X-linked |
2191 |
0.25 |
| chr4_123904447_123904800 | 0.46 |
Mycbp |
MYC binding protein |
209 |
0.77 |
| chr6_16316653_16317675 | 0.46 |
Gm3148 |
predicted gene 3148 |
79537 |
0.1 |
| chr2_76980555_76980961 | 0.45 |
Ttn |
titin |
576 |
0.81 |
| chr7_93079849_93081191 | 0.45 |
Gm9934 |
predicted gene 9934 |
274 |
0.76 |
| chr11_57836623_57837461 | 0.45 |
Hand1 |
heart and neural crest derivatives expressed 1 |
4224 |
0.18 |
| chr4_150807641_150808455 | 0.45 |
Gm13049 |
predicted gene 13049 |
17685 |
0.16 |
| chr1_40807253_40807432 | 0.44 |
Tmem182 |
transmembrane protein 182 |
1741 |
0.33 |
| chr11_5802324_5803834 | 0.44 |
Pgam2 |
phosphoglycerate mutase 2 |
654 |
0.56 |
| chr1_72826047_72827238 | 0.44 |
Igfbp2 |
insulin-like growth factor binding protein 2 |
1320 |
0.5 |
| chr7_99350039_99350190 | 0.44 |
Serpinh1 |
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
2883 |
0.2 |
| chr17_17827521_17828536 | 0.44 |
Spaca6 |
sperm acrosome associated 6 |
704 |
0.43 |
| chr4_110050529_110051058 | 0.44 |
Dmrta2 |
doublesex and mab-3 related transcription factor like family A2 |
72740 |
0.11 |
| chr4_119463634_119463785 | 0.44 |
Zmynd12 |
zinc finger, MYND domain containing 12 |
18989 |
0.09 |
| chr2_150788802_150789695 | 0.44 |
Pygb |
brain glycogen phosphorylase |
242 |
0.91 |
| chr18_49559060_49559779 | 0.43 |
Gm25440 |
predicted gene, 25440 |
22430 |
0.22 |
| chr7_142474137_142474301 | 0.43 |
Lsp1 |
lymphocyte specific 1 |
775 |
0.47 |
| chr12_29665011_29665344 | 0.43 |
C630031E19Rik |
RIKEN cDNA C630031E19 gene |
21268 |
0.26 |
| chr5_104109155_104109917 | 0.43 |
Gm26703 |
predicted gene, 26703 |
245 |
0.88 |
| chr11_94498570_94500249 | 0.43 |
Epn3 |
epsin 3 |
289 |
0.85 |
| chr7_70338252_70338403 | 0.43 |
Gm29683 |
predicted gene, 29683 |
475 |
0.72 |
| chr11_6604576_6606131 | 0.43 |
Nacad |
NAC alpha domain containing |
700 |
0.46 |
| chrY_1041845_1042254 | 0.42 |
Gm29650 |
predicted gene 29650 |
6344 |
0.16 |
| chr13_12104357_12104508 | 0.42 |
Ryr2 |
ryanodine receptor 2, cardiac |
2027 |
0.32 |
| chr10_57001100_57001565 | 0.42 |
Gm36827 |
predicted gene, 36827 |
19100 |
0.24 |
| chr4_108830914_108831550 | 0.42 |
Btf3l4 |
basic transcription factor 3-like 4 |
681 |
0.59 |
| chr2_178118695_178119817 | 0.42 |
Phactr3 |
phosphatase and actin regulator 3 |
77 |
0.98 |
| chr13_47770992_47771224 | 0.42 |
4930471G24Rik |
RIKEN cDNA 4930471G24 gene |
103558 |
0.07 |
| chr4_151137762_151138535 | 0.42 |
Camta1 |
calmodulin binding transcription activator 1 |
1436 |
0.47 |
| chr17_6772344_6772495 | 0.41 |
Ezr |
ezrin |
10365 |
0.16 |
| chr15_27786588_27788615 | 0.41 |
Trio |
triple functional domain (PTPRF interacting) |
1037 |
0.6 |
| chr7_29066413_29066564 | 0.41 |
Gm26604 |
predicted gene, 26604 |
5115 |
0.1 |
| chr18_37178155_37179152 | 0.41 |
Gm10544 |
predicted gene 10544 |
131 |
0.94 |
| chr17_17448568_17449744 | 0.41 |
Lix1 |
limb and CNS expressed 1 |
336 |
0.88 |
| chr14_45850711_45850862 | 0.41 |
Gm8317 |
predicted gene 8317 |
7760 |
0.2 |
| chr7_46407345_46407496 | 0.40 |
Kcnc1 |
potassium voltage gated channel, Shaw-related subfamily, member 1 |
9772 |
0.16 |
| chr1_57573806_57574059 | 0.40 |
Gm17929 |
predicted gene, 17929 |
9881 |
0.21 |
| chr15_82277282_82277951 | 0.40 |
Septin3 |
septin 3 |
2216 |
0.14 |
| chr11_21570140_21570702 | 0.40 |
Mdh1 |
malate dehydrogenase 1, NAD (soluble) |
1513 |
0.29 |
| chr4_129141555_129141750 | 0.40 |
Fndc5 |
fibronectin type III domain containing 5 |
4653 |
0.13 |
| chr2_26339789_26340241 | 0.40 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
567 |
0.56 |
| chr8_11309069_11309952 | 0.40 |
Col4a1 |
collagen, type IV, alpha 1 |
3179 |
0.2 |
| chr7_4516649_4516857 | 0.40 |
Tnnt1 |
troponin T1, skeletal, slow |
371 |
0.69 |
| chr1_118481967_118482873 | 0.39 |
Clasp1 |
CLIP associating protein 1 |
381 |
0.8 |
| chr17_57058009_57058459 | 0.39 |
Crb3 |
crumbs family member 3 |
871 |
0.3 |
| chr9_99662299_99662551 | 0.39 |
Dzip1l |
DAZ interacting protein 1-like |
12209 |
0.16 |
| chr11_94045498_94046076 | 0.39 |
Spag9 |
sperm associated antigen 9 |
1422 |
0.4 |
| chr6_137168523_137169703 | 0.39 |
Rerg |
RAS-like, estrogen-regulated, growth-inhibitor |
572 |
0.79 |
| chr10_98967523_98967758 | 0.39 |
Atp2b1 |
ATPase, Ca++ transporting, plasma membrane 1 |
18491 |
0.22 |
| chr15_77021062_77022444 | 0.39 |
Mb |
myoglobin |
210 |
0.89 |
| chr16_84549281_84549553 | 0.39 |
A730009L09Rik |
RIKEN cDNA A730009L09 gene |
34048 |
0.14 |
| chr18_42444794_42444945 | 0.39 |
Gm16415 |
predicted pseudogene 16415 |
3850 |
0.22 |
| chr14_55057067_55057894 | 0.39 |
Gm20687 |
predicted gene 20687 |
1987 |
0.14 |
| chr14_60383740_60384237 | 0.39 |
Amer2 |
APC membrane recruitment 2 |
5702 |
0.23 |
| chr2_163379610_163379927 | 0.39 |
Jph2 |
junctophilin 2 |
18181 |
0.13 |
| chr2_13578293_13579079 | 0.38 |
Vim |
vimentin |
683 |
0.75 |
| chr3_18049845_18050841 | 0.38 |
Bhlhe22 |
basic helix-loop-helix family, member e22 |
3831 |
0.23 |
| chr4_108218802_108219449 | 0.38 |
Zyg11a |
zyg-11 family member A, cell cycle regulator |
1077 |
0.46 |
| chr13_110018579_110019682 | 0.38 |
Pde4d |
phosphodiesterase 4D, cAMP specific |
83818 |
0.1 |
| chr7_99467468_99467619 | 0.38 |
Klhl35 |
kelch-like 35 |
1420 |
0.27 |
| chr10_95894136_95895107 | 0.37 |
Gm47725 |
predicted gene, 47725 |
1178 |
0.38 |
| chr2_75086892_75087412 | 0.37 |
Gm13652 |
predicted gene 13652 |
50646 |
0.11 |
| chr19_53679904_53680339 | 0.37 |
Rbm20 |
RNA binding motif protein 20 |
2815 |
0.26 |
| chr3_75921070_75921248 | 0.37 |
Golim4 |
golgi integral membrane protein 4 |
18713 |
0.18 |
| chr8_3606652_3606803 | 0.37 |
Camsap3 |
calmodulin regulated spectrin-associated protein family, member 3 |
389 |
0.68 |
| chr17_47975965_47976586 | 0.37 |
Gm14871 |
predicted gene 14871 |
27297 |
0.11 |
| chr6_25688825_25690399 | 0.37 |
Gpr37 |
G protein-coupled receptor 37 |
180 |
0.97 |
| chr2_173270600_173271108 | 0.37 |
Pmepa1 |
prostate transmembrane protein, androgen induced 1 |
5335 |
0.2 |
| chr3_102086617_102087454 | 0.36 |
Casq2 |
calsequestrin 2 |
396 |
0.81 |
| chr3_134330789_134331989 | 0.36 |
Gm43558 |
predicted gene 43558 |
25903 |
0.15 |
| chrX_143929366_143930062 | 0.36 |
Dcx |
doublecortin |
3336 |
0.34 |
| chr6_6212569_6212720 | 0.36 |
Slc25a13 |
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
4474 |
0.27 |
| chr2_49787205_49788356 | 0.36 |
Lypd6b |
LY6/PLAUR domain containing 6B |
49 |
0.98 |
| chr17_25462833_25463318 | 0.36 |
Tekt4 |
tektin 4 |
8460 |
0.11 |
| chr15_103506702_103507198 | 0.35 |
Pde1b |
phosphodiesterase 1B, Ca2+-calmodulin dependent |
3673 |
0.17 |
| chr13_15550714_15550912 | 0.35 |
Gli3 |
GLI-Kruppel family member GLI3 |
86833 |
0.07 |
| chr10_80940377_80940528 | 0.35 |
Gadd45b |
growth arrest and DNA-damage-inducible 45 beta |
9640 |
0.09 |
| chr16_20699849_20700000 | 0.35 |
Fam131a |
family with sequence similarity 131, member A |
3749 |
0.09 |
| chr2_153035186_153035549 | 0.35 |
Xkr7 |
X-linked Kx blood group related 7 |
3515 |
0.16 |
| chr9_58824792_58824943 | 0.35 |
Hcn4 |
hyperpolarization-activated, cyclic nucleotide-gated K+ 4 |
1455 |
0.49 |
| chr17_78734767_78735199 | 0.35 |
Strn |
striatin, calmodulin binding protein |
2213 |
0.24 |
| chr1_131522570_131523013 | 0.34 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
4556 |
0.17 |
| chr14_111680945_111682200 | 0.34 |
Slitrk5 |
SLIT and NTRK-like family, member 5 |
5723 |
0.23 |
| chr19_5474276_5474674 | 0.34 |
Efemp2 |
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
160 |
0.83 |
| chr3_87996506_87997409 | 0.34 |
Bcan |
brevican |
308 |
0.8 |
| chr10_80296711_80297037 | 0.34 |
Apc2 |
APC regulator of WNT signaling pathway 2 |
897 |
0.3 |
| chr5_101365618_101366080 | 0.34 |
Cycs-ps2 |
cytochrome c, pseudogene 2 |
60271 |
0.14 |
| chr11_95521077_95521771 | 0.34 |
Nxph3 |
neurexophilin 3 |
6854 |
0.18 |
| chr9_47671566_47671717 | 0.34 |
Gm47198 |
predicted gene, 47198 |
99500 |
0.07 |
| chr7_25069270_25070477 | 0.34 |
Grik5 |
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
2228 |
0.18 |
| chr2_160871307_160871738 | 0.33 |
Zhx3 |
zinc fingers and homeoboxes 3 |
1050 |
0.42 |
| chr13_48624936_48625515 | 0.33 |
Ptpdc1 |
protein tyrosine phosphatase domain containing 1 |
284 |
0.88 |
| chr8_27263616_27263874 | 0.33 |
Eif4ebp1 |
eukaryotic translation initiation factor 4E binding protein 1 |
669 |
0.56 |
| chr9_118480279_118480840 | 0.33 |
Eomes |
eomesodermin |
1698 |
0.28 |
| chr17_24552226_24552377 | 0.33 |
Pkd1 |
polycystin 1, transient receptor poteintial channel interacting |
2351 |
0.12 |
| chr14_58113076_58113458 | 0.33 |
Gm17109 |
predicted gene 17109 |
7138 |
0.21 |
| chr9_52052762_52052932 | 0.33 |
Rdx |
radixin |
4576 |
0.21 |
| chr3_57625355_57625848 | 0.33 |
Gm26671 |
predicted gene, 26671 |
156 |
0.92 |
| chr5_103209022_103210413 | 0.32 |
Mapk10 |
mitogen-activated protein kinase 10 |
705 |
0.75 |
| chr14_58150040_58150734 | 0.32 |
Gm17109 |
predicted gene 17109 |
44258 |
0.14 |
| chr17_34185809_34185960 | 0.32 |
Psmb9 |
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
1335 |
0.17 |
| chr14_8665553_8666279 | 0.32 |
4930452B06Rik |
RIKEN cDNA 4930452B06 gene |
323 |
0.62 |
| chr8_45673773_45673924 | 0.32 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
11852 |
0.21 |
| chr16_85091082_85091583 | 0.31 |
Gm49227 |
predicted gene, 49227 |
11221 |
0.2 |
| chr5_41766502_41767052 | 0.31 |
Nkx3-2 |
NK3 homeobox 2 |
2276 |
0.35 |
| chr1_187653088_187653646 | 0.31 |
Gm36388 |
predicted gene, 36388 |
1543 |
0.4 |
| chr4_141964364_141964893 | 0.31 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
10434 |
0.13 |
| chr10_92159283_92159475 | 0.31 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
3382 |
0.26 |
| chr4_105449104_105449638 | 0.31 |
Gm12723 |
predicted gene 12723 |
61861 |
0.13 |
| chr1_83407298_83408547 | 0.31 |
Sphkap |
SPHK1 interactor, AKAP domain containing |
217 |
0.94 |
| chr7_45240555_45241594 | 0.31 |
Slc6a16 |
solute carrier family 6, member 16 |
11 |
0.92 |
| chr10_128896056_128896905 | 0.30 |
Cd63 |
CD63 antigen |
4509 |
0.1 |
| chr14_66792058_66792209 | 0.30 |
Gm9130 |
predicted gene 9130 |
6245 |
0.19 |
| chr10_69536011_69536410 | 0.30 |
Ank3 |
ankyrin 3, epithelial |
1988 |
0.39 |
| chr8_79030967_79031125 | 0.30 |
Zfp827 |
zinc finger protein 827 |
2459 |
0.3 |
| chr6_87432429_87432774 | 0.30 |
Bmp10 |
bone morphogenetic protein 10 |
3607 |
0.18 |
| chr14_58072231_58072956 | 0.30 |
Fgf9 |
fibroblast growth factor 9 |
93 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 0.8 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.2 | 1.2 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 0.6 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 0.5 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.2 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.5 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.1 | 0.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 0.2 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.1 | 0.2 | GO:1900238 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.4 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.3 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.2 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.1 | 0.4 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.2 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.1 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.1 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.1 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.0 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 1.8 | GO:0055010 | ventricular cardiac muscle tissue morphogenesis(GO:0055010) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:1903242 | regulation of cardiac muscle adaptation(GO:0010612) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.0 | GO:2001182 | regulation of interleukin-12 secretion(GO:2001182) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.0 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0097012 | response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.0 | GO:1901321 | positive regulation of heart induction(GO:1901321) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.2 | GO:0046218 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.0 | GO:2000850 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.0 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0051794 | regulation of catagen(GO:0051794) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:2001201 | transforming growth factor-beta secretion(GO:0038044) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.2 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.0 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.3 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0018119 | peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.0 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0086029 | Purkinje myocyte action potential(GO:0086017) Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0043307 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.1 | GO:0071071 | regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.0 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.0 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.0 | 0.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.0 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:1903660 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.2 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.0 | GO:1902302 | potassium ion export across plasma membrane(GO:0097623) regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.5 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 3.8 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 0.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 0.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 0.5 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.0 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0052623 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0043726 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |