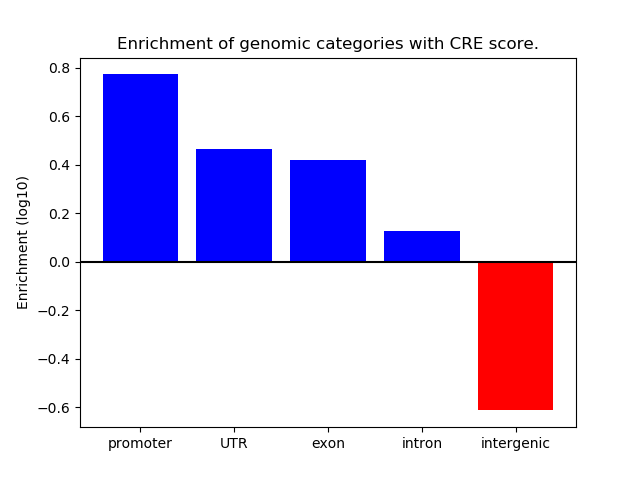
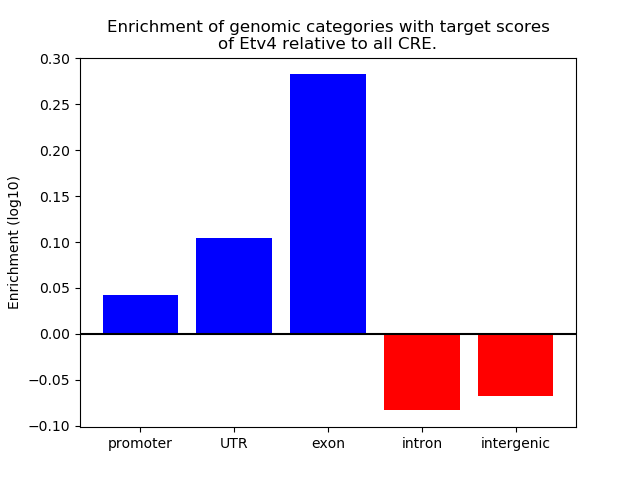
Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
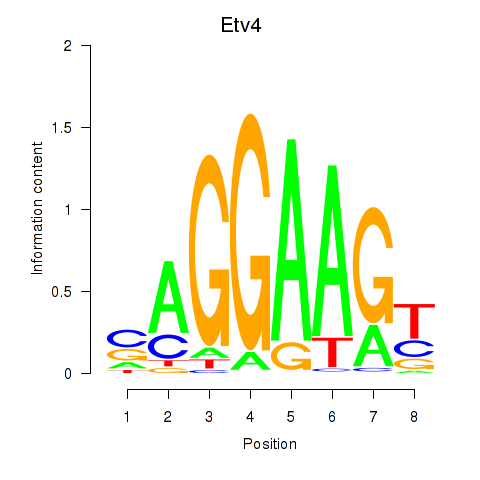
Results for Etv4
Z-value: 0.44
Transcription factors associated with Etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv4
|
ENSMUSG00000017724.8 | ets variant 4 |
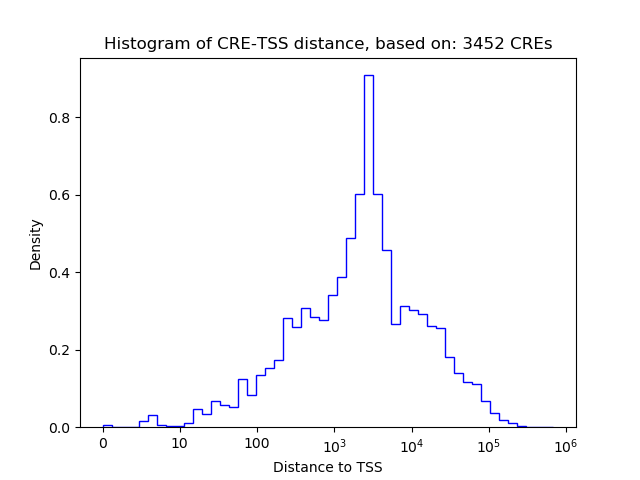
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_101814398_101814872 | Etv4 | 29264 | 0.120665 | 0.49 | 6.8e-05 | Click! |
| chr11_101784179_101785383 | Etv4 | 67 | 0.967360 | 0.35 | 6.0e-03 | Click! |
| chr11_101782309_101782733 | Etv4 | 1867 | 0.286426 | 0.35 | 6.1e-03 | Click! |
| chr11_101814893_101815371 | Etv4 | 29761 | 0.119678 | 0.33 | 9.3e-03 | Click! |
| chr11_101806597_101806748 | Etv4 | 21301 | 0.135767 | 0.31 | 1.6e-02 | Click! |
Activity of the Etv4 motif across conditions
Conditions sorted by the z-value of the Etv4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
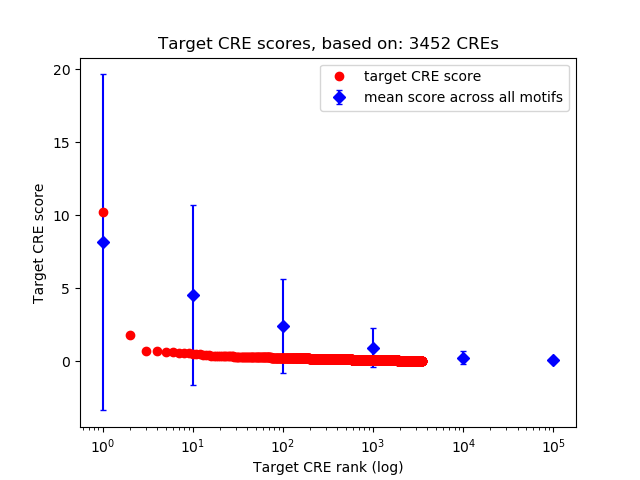
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_170674573_170675954 | 10.21 |
Asmt |
acetylserotonin O-methyltransferase |
2619 |
0.41 |
| chr7_4518554_4518985 | 1.83 |
Tnni3 |
troponin I, cardiac 3 |
995 |
0.29 |
| chr12_3236855_3237648 | 0.72 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
640 |
0.66 |
| chr9_124439879_124441093 | 0.71 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
382 |
0.83 |
| chr14_106163700_106163851 | 0.66 |
Gm48971 |
predicted gene, 48971 |
48203 |
0.12 |
| chr5_112215410_112216373 | 0.64 |
Gm26953 |
predicted gene, 26953 |
430 |
0.76 |
| chr8_45509543_45510137 | 0.56 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
1922 |
0.33 |
| chr11_94045498_94046076 | 0.54 |
Spag9 |
sperm associated antigen 9 |
1422 |
0.4 |
| chr4_89285556_89285707 | 0.54 |
Cdkn2a |
cyclin dependent kinase inhibitor 2A |
3439 |
0.19 |
| chr2_71525880_71527745 | 0.51 |
Dlx1as |
distal-less homeobox 1, antisense |
1107 |
0.33 |
| chr13_83721518_83722206 | 0.51 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
481 |
0.73 |
| chr1_133735425_133736534 | 0.50 |
Mir6903 |
microRNA 6903 |
9334 |
0.14 |
| chr10_29143400_29144848 | 0.44 |
Soga3 |
SOGA family member 3 |
65 |
0.5 |
| chr3_30649995_30651146 | 0.44 |
Lrriq4 |
leucine-rich repeats and IQ motif containing 4 |
2684 |
0.17 |
| chr1_184235265_184236073 | 0.41 |
Gm37223 |
predicted gene, 37223 |
122660 |
0.05 |
| chr4_45824847_45825701 | 0.40 |
Igfbpl1 |
insulin-like growth factor binding protein-like 1 |
1649 |
0.31 |
| chr12_29529828_29531185 | 0.38 |
Gm20208 |
predicted gene, 20208 |
609 |
0.74 |
| chr3_108410436_108412210 | 0.38 |
Celsr2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
4229 |
0.11 |
| chr2_140666876_140668088 | 0.37 |
Flrt3 |
fibronectin leucine rich transmembrane protein 3 |
3918 |
0.37 |
| chr13_112695809_112696082 | 0.37 |
Gm18883 |
predicted gene, 18883 |
3734 |
0.17 |
| chr16_10446425_10446612 | 0.37 |
Tvp23a |
trans-golgi network vesicle protein 23A |
626 |
0.68 |
| chr12_29528407_29529244 | 0.36 |
Myt1l |
myelin transcription factor 1-like |
424 |
0.85 |
| chr4_89285170_89285321 | 0.35 |
Cdkn2a |
cyclin dependent kinase inhibitor 2A |
3053 |
0.2 |
| chr9_35423074_35423780 | 0.34 |
Cdon |
cell adhesion molecule-related/down-regulated by oncogenes |
161 |
0.95 |
| chr7_25294533_25294696 | 0.34 |
Cic |
capicua transcriptional repressor |
1594 |
0.18 |
| chr14_122475443_122476757 | 0.34 |
Zic2 |
zinc finger protein of the cerebellum 2 |
665 |
0.44 |
| chr7_44441951_44442938 | 0.33 |
Lrrc4b |
leucine rich repeat containing 4B |
41 |
0.93 |
| chr3_34656299_34657721 | 0.33 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
974 |
0.4 |
| chr1_42691569_42692627 | 0.33 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
995 |
0.42 |
| chr3_94479508_94480443 | 0.33 |
Riiad1 |
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
366 |
0.63 |
| chr12_12263781_12264571 | 0.32 |
Fam49a |
family with sequence similarity 49, member A |
1987 |
0.46 |
| chr12_117157079_117158175 | 0.32 |
Gm10421 |
predicted gene 10421 |
5976 |
0.31 |
| chr7_4920681_4922210 | 0.32 |
Nat14 |
N-acetyltransferase 14 |
583 |
0.48 |
| chr17_46617075_46618033 | 0.32 |
Ptk7 |
PTK7 protein tyrosine kinase 7 |
11950 |
0.1 |
| chrX_123943318_123943469 | 0.32 |
Rps12-ps20 |
ribosomal protein S12, pseudogene 20 |
13881 |
0.22 |
| chr1_5916517_5917959 | 0.32 |
Npbwr1 |
neuropeptides B/W receptor 1 |
160 |
0.97 |
| chr4_156183474_156184054 | 0.31 |
Agrn |
agrin |
2137 |
0.17 |
| chr1_115684558_115685809 | 0.31 |
Cntnap5a |
contactin associated protein-like 5A |
427 |
0.58 |
| chr6_121472423_121472626 | 0.31 |
Iqsec3 |
IQ motif and Sec7 domain 3 |
1099 |
0.53 |
| chr5_120433178_120434996 | 0.31 |
Gm27199 |
predicted gene 27199 |
2320 |
0.19 |
| chr12_33928070_33929277 | 0.31 |
Gm40383 |
predicted gene, 40383 |
217 |
0.53 |
| chr4_96552598_96553907 | 0.31 |
Cyp2j6 |
cytochrome P450, family 2, subfamily j, polypeptide 6 |
409 |
0.87 |
| chr8_94994139_94995207 | 0.30 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
77 |
0.95 |
| chr3_114904046_114905354 | 0.30 |
Olfm3 |
olfactomedin 3 |
65 |
0.98 |
| chr9_29943252_29944148 | 0.30 |
Ntm |
neurotrimin |
19419 |
0.28 |
| chr10_80298461_80300404 | 0.30 |
Apc2 |
APC regulator of WNT signaling pathway 2 |
341 |
0.69 |
| chr12_65428156_65429051 | 0.30 |
Gm26015 |
predicted gene, 26015 |
22633 |
0.22 |
| chr3_26332588_26333347 | 0.29 |
Nlgn1 |
neuroligin 1 |
507 |
0.68 |
| chr6_13835523_13837039 | 0.29 |
Gpr85 |
G protein-coupled receptor 85 |
960 |
0.59 |
| chr15_95525022_95525792 | 0.29 |
Nell2 |
NEL-like 2 |
2771 |
0.35 |
| chr6_90781027_90782541 | 0.29 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
188 |
0.94 |
| chr4_48586042_48586294 | 0.29 |
Tmeff1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
617 |
0.75 |
| chr7_126978893_126979102 | 0.28 |
Cdipt |
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
764 |
0.32 |
| chr8_41052368_41053980 | 0.28 |
Gm16193 |
predicted gene 16193 |
64 |
0.96 |
| chr5_138278285_138278436 | 0.28 |
Gpc2 |
glypican 2 (cerebroglycan) |
123 |
0.87 |
| chr16_96280491_96281572 | 0.28 |
B3galt5 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
233 |
0.93 |
| chr14_76420544_76421824 | 0.27 |
Tsc22d1 |
TSC22 domain family, member 1 |
2356 |
0.39 |
| chr18_15060980_15061427 | 0.27 |
Kctd1 |
potassium channel tetramerisation domain containing 1 |
686 |
0.74 |
| chr9_97017174_97017806 | 0.27 |
Gm16010 |
predicted gene 16010 |
16 |
0.96 |
| chr13_51566607_51568077 | 0.27 |
Shc3 |
src homology 2 domain-containing transforming protein C3 |
258 |
0.94 |
| chr4_13746200_13747057 | 0.27 |
Runx1t1 |
RUNX1 translocation partner 1 |
3192 |
0.37 |
| chr9_108824114_108825614 | 0.27 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr12_3370750_3371679 | 0.27 |
Gm48511 |
predicted gene, 48511 |
2785 |
0.18 |
| chr1_6733530_6734383 | 0.27 |
St18 |
suppression of tumorigenicity 18 |
914 |
0.7 |
| chr7_78884387_78884983 | 0.27 |
Mir7-2 |
microRNA 7-2 |
3592 |
0.14 |
| chr10_60115211_60116020 | 0.27 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
9147 |
0.19 |
| chr4_15880938_15882113 | 0.26 |
Calb1 |
calbindin 1 |
259 |
0.9 |
| chr10_127268122_127269422 | 0.26 |
Dctn2 |
dynactin 2 |
2385 |
0.12 |
| chr16_9994378_9995594 | 0.26 |
Grin2a |
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
63 |
0.98 |
| chr2_166152712_166153273 | 0.26 |
Sulf2 |
sulfatase 2 |
1431 |
0.4 |
| chr5_34182980_34183403 | 0.26 |
Mxd4 |
Max dimerization protein 4 |
1179 |
0.3 |
| chr2_51144318_51145477 | 0.26 |
Rnd3 |
Rho family GTPase 3 |
4197 |
0.29 |
| chr1_104768492_104769666 | 0.26 |
Cdh20 |
cadherin 20 |
550 |
0.79 |
| chr3_34197711_34199105 | 0.26 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
1544 |
0.41 |
| chr7_10480473_10481196 | 0.25 |
Gm30645 |
predicted gene, 30645 |
11141 |
0.11 |
| chr5_148265188_148266164 | 0.25 |
Mtus2 |
microtubule associated tumor suppressor candidate 2 |
331 |
0.91 |
| chr14_14345697_14346878 | 0.25 |
Il3ra |
interleukin 3 receptor, alpha chain |
12 |
0.94 |
| chr11_69043818_69044451 | 0.25 |
Aurkb |
aurora kinase B |
1513 |
0.17 |
| chr7_126277403_126277977 | 0.25 |
Sbk1 |
SH3-binding kinase 1 |
4290 |
0.14 |
| chr10_3863416_3864702 | 0.25 |
Gm16149 |
predicted gene 16149 |
5548 |
0.21 |
| chr19_4758474_4759201 | 0.25 |
Rbm4b |
RNA binding motif protein 4B |
1932 |
0.18 |
| chr11_76466993_76468433 | 0.25 |
Abr |
active BCR-related gene |
585 |
0.77 |
| chr11_57013425_57013750 | 0.25 |
Gria1 |
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
541 |
0.88 |
| chr14_79773410_79774519 | 0.25 |
Pcdh8 |
protocadherin 8 |
2652 |
0.21 |
| chr10_59950302_59950479 | 0.25 |
Ddit4 |
DNA-damage-inducible transcript 4 |
1444 |
0.4 |
| chr2_20339506_20340619 | 0.25 |
Etl4 |
enhancer trap locus 4 |
238 |
0.94 |
| chr8_121300864_121302289 | 0.24 |
Gm26815 |
predicted gene, 26815 |
55260 |
0.11 |
| chr9_52145956_52147029 | 0.24 |
Zc3h12c |
zinc finger CCCH type containing 12C |
21619 |
0.17 |
| chr2_65929929_65930575 | 0.24 |
Csrnp3 |
cysteine-serine-rich nuclear protein 3 |
115 |
0.97 |
| chr4_82499658_82501360 | 0.24 |
Nfib |
nuclear factor I/B |
1193 |
0.5 |
| chr12_117149811_117150131 | 0.24 |
Gm10421 |
predicted gene 10421 |
1244 |
0.61 |
| chr13_51594475_51595717 | 0.24 |
Shc3 |
src homology 2 domain-containing transforming protein C3 |
25609 |
0.16 |
| chr4_22835212_22836357 | 0.24 |
Gm24078 |
predicted gene, 24078 |
88653 |
0.09 |
| chr14_14351494_14351697 | 0.24 |
Il3ra |
interleukin 3 receptor, alpha chain |
1974 |
0.2 |
| chr15_92160888_92162023 | 0.24 |
Cntn1 |
contactin 1 |
98 |
0.98 |
| chr14_68126289_68127413 | 0.23 |
Nefm |
neurofilament, medium polypeptide |
2005 |
0.3 |
| chr7_133757893_133758428 | 0.23 |
Dhx32 |
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
1312 |
0.34 |
| chr1_25832036_25832712 | 0.23 |
Gm9884 |
predicted gene 9884 |
1717 |
0.22 |
| chr18_15060481_15060964 | 0.23 |
Kctd1 |
potassium channel tetramerisation domain containing 1 |
1167 |
0.55 |
| chr7_67421454_67422158 | 0.23 |
Gm33926 |
predicted gene, 33926 |
22127 |
0.17 |
| chr9_47533891_47534042 | 0.23 |
Cadm1 |
cell adhesion molecule 1 |
3593 |
0.24 |
| chr11_64520049_64520796 | 0.23 |
Gm24275 |
predicted gene, 24275 |
70218 |
0.12 |
| chr2_71536695_71536971 | 0.23 |
Dlx1as |
distal-less homeobox 1, antisense |
977 |
0.45 |
| chr1_34677886_34678995 | 0.23 |
Arhgef4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
252 |
0.89 |
| chr2_167775422_167775573 | 0.23 |
Gm14321 |
predicted gene 14321 |
1970 |
0.27 |
| chr2_80128704_80129752 | 0.23 |
Pde1a |
phosphodiesterase 1A, calmodulin-dependent |
230 |
0.94 |
| chr14_28508967_28511864 | 0.23 |
Wnt5a |
wingless-type MMTV integration site family, member 5A |
203 |
0.89 |
| chr3_31097203_31098928 | 0.23 |
Skil |
SKI-like |
1232 |
0.48 |
| chr11_71749802_71749953 | 0.23 |
Wscd1 |
WSC domain containing 1 |
43 |
0.97 |
| chr1_77503797_77505059 | 0.23 |
Mir6352 |
microRNA 6352 |
7660 |
0.19 |
| chr7_81494043_81494194 | 0.23 |
Ap3b2 |
adaptor-related protein complex 3, beta 2 subunit |
193 |
0.89 |
| chr5_28466220_28467111 | 0.23 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
319 |
0.7 |
| chr16_14906219_14907566 | 0.23 |
Efcab1 |
EF-hand calcium binding domain 1 |
227 |
0.95 |
| chr4_41693576_41694021 | 0.23 |
Cntfr |
ciliary neurotrophic factor receptor |
1644 |
0.21 |
| chr14_12339117_12339550 | 0.23 |
Gm24578 |
predicted gene, 24578 |
5429 |
0.13 |
| chr13_105249326_105250833 | 0.23 |
Rnf180 |
ring finger protein 180 |
20960 |
0.22 |
| chr12_5374545_5374864 | 0.23 |
Klhl29 |
kelch-like 29 |
978 |
0.45 |
| chr10_19358667_19359238 | 0.23 |
Olig3 |
oligodendrocyte transcription factor 3 |
2419 |
0.35 |
| chr4_72203288_72203508 | 0.22 |
C630043F03Rik |
RIKEN cDNA C630043F03 gene |
2050 |
0.31 |
| chr17_35160468_35162406 | 0.22 |
Gm23442 |
predicted gene, 23442 |
1259 |
0.13 |
| chr7_87586513_87587584 | 0.22 |
Grm5 |
glutamate receptor, metabotropic 5 |
2650 |
0.4 |
| chr11_113621749_113621900 | 0.22 |
Sstr2 |
somatostatin receptor 2 |
2348 |
0.25 |
| chr3_83768357_83769273 | 0.22 |
Sfrp2 |
secreted frizzled-related protein 2 |
1962 |
0.31 |
| chr11_103776117_103777283 | 0.22 |
Wnt3 |
wingless-type MMTV integration site family, member 3 |
2550 |
0.24 |
| chr7_16948034_16948663 | 0.22 |
Pnmal2 |
PNMA-like 2 |
3666 |
0.11 |
| chr2_129908592_129909805 | 0.22 |
Gm28196 |
predicted gene 28196 |
39370 |
0.15 |
| chr2_155514247_155515194 | 0.22 |
Ggt7 |
gamma-glutamyltransferase 7 |
128 |
0.92 |
| chr11_88584998_88585279 | 0.22 |
Msi2 |
musashi RNA-binding protein 2 |
5009 |
0.28 |
| chr8_109245493_109246323 | 0.22 |
D030068K23Rik |
RIKEN cDNA D030068K23 gene |
3958 |
0.33 |
| chr1_131523488_131524457 | 0.22 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
3375 |
0.18 |
| chr3_55782570_55784448 | 0.22 |
Nbea |
neurobeachin |
19 |
0.96 |
| chr12_27140199_27140495 | 0.22 |
Gm9866 |
predicted gene 9866 |
19964 |
0.27 |
| chr17_93201490_93204144 | 0.22 |
Adcyap1 |
adenylate cyclase activating polypeptide 1 |
741 |
0.65 |
| chr17_28256852_28257467 | 0.22 |
Ppard |
peroxisome proliferator activator receptor delta |
14549 |
0.11 |
| chr1_166256819_166257273 | 0.22 |
Ildr2 |
immunoglobulin-like domain containing receptor 2 |
2853 |
0.25 |
| chr12_91683421_91686419 | 0.22 |
Gm16876 |
predicted gene, 16876 |
255 |
0.86 |
| chr5_98182267_98183697 | 0.21 |
Prdm8 |
PR domain containing 8 |
2004 |
0.26 |
| chr3_87539034_87539185 | 0.21 |
ETV3L |
ets variant 3-like |
10990 |
0.16 |
| chrX_134405492_134405850 | 0.21 |
Drp2 |
dystrophin related protein 2 |
869 |
0.59 |
| chr7_79514267_79514939 | 0.21 |
Mir9-3hg |
Mir9-3 host gene |
629 |
0.46 |
| chr5_5507730_5508026 | 0.21 |
Cldn12 |
claudin 12 |
6571 |
0.17 |
| chr15_74563319_74564610 | 0.21 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
63 |
0.61 |
| chrX_101708550_101709190 | 0.21 |
Gm9089 |
predicted gene 9089 |
691 |
0.52 |
| chr9_78377654_78378912 | 0.21 |
Ooep |
oocyte expressed protein |
442 |
0.65 |
| chr5_66967831_66968776 | 0.21 |
Limch1 |
LIM and calponin homology domains 1 |
113 |
0.93 |
| chr8_58912073_58913264 | 0.21 |
Galntl6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
28 |
0.88 |
| chr2_120848173_120848373 | 0.21 |
Cdan1 |
congenital dyserythropoietic anemia, type I (human) |
1855 |
0.22 |
| chrX_96712746_96713886 | 0.21 |
Gpr165 |
G protein-coupled receptor 165 |
125 |
0.98 |
| chr17_52600782_52601825 | 0.21 |
Gm27217 |
predicted gene 27217 |
1357 |
0.38 |
| chr1_119420327_119420935 | 0.21 |
Inhbb |
inhibin beta-B |
1617 |
0.38 |
| chr9_102721179_102722320 | 0.21 |
Amotl2 |
angiomotin-like 2 |
567 |
0.65 |
| chr4_156182317_156182745 | 0.21 |
Agrn |
agrin |
3370 |
0.13 |
| chr4_99118711_99119482 | 0.20 |
Dock7 |
dedicator of cytokinesis 7 |
1666 |
0.29 |
| chr4_141998561_141998861 | 0.20 |
Fhad1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
12926 |
0.12 |
| chr9_48517322_48517969 | 0.20 |
Gm23653 |
predicted gene, 23653 |
10585 |
0.15 |
| chrX_74310114_74310288 | 0.20 |
Gdi1 |
guanosine diphosphate (GDP) dissociation inhibitor 1 |
180 |
0.84 |
| chr4_72195085_72196381 | 0.20 |
Tle1 |
transducin-like enhancer of split 1 |
3359 |
0.26 |
| chr7_90886512_90887836 | 0.20 |
Gm45159 |
predicted gene 45159 |
102 |
0.88 |
| chr16_22160432_22161001 | 0.20 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
734 |
0.66 |
| chr14_13283913_13285295 | 0.20 |
Gm5087 |
predicted gene 5087 |
26 |
0.69 |
| chr2_151966993_151968089 | 0.20 |
Mir1953 |
microRNA 1953 |
76 |
0.96 |
| chr9_87727084_87728023 | 0.20 |
D030062O11Rik |
RIKEN cDNA D030062O11 gene |
1732 |
0.31 |
| chr9_29411568_29412757 | 0.20 |
Ntm |
neurotrimin |
42 |
0.99 |
| chr15_100872381_100872532 | 0.20 |
Scn8a |
sodium channel, voltage-gated, type VIII, alpha |
1036 |
0.52 |
| chr9_121493633_121494845 | 0.20 |
Cck |
cholecystokinin |
271 |
0.9 |
| chr18_16804620_16805078 | 0.20 |
Cdh2 |
cadherin 2 |
3613 |
0.25 |
| chr4_123001785_123002093 | 0.20 |
Mycl |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived |
2600 |
0.21 |
| chr12_3368013_3368276 | 0.20 |
Gm48511 |
predicted gene, 48511 |
285 |
0.85 |
| chr2_38824726_38825619 | 0.20 |
Nr6a1os |
nuclear receptor subfamily 6, group A, member 1, opposite strand |
1831 |
0.22 |
| chr19_55898749_55899177 | 0.20 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
654 |
0.79 |
| chr3_89523444_89523616 | 0.20 |
Kcnn3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
3366 |
0.2 |
| chr1_56969244_56970464 | 0.20 |
Satb2 |
special AT-rich sequence binding protein 2 |
14 |
0.97 |
| chr12_27334769_27335068 | 0.20 |
Sox11 |
SRY (sex determining region Y)-box 11 |
7656 |
0.29 |
| chr17_8592901_8593278 | 0.20 |
Gm15425 |
predicted gene 15425 |
24229 |
0.18 |
| chr8_70691465_70691881 | 0.20 |
Jund |
jun D proto-oncogene |
7276 |
0.08 |
| chr15_101995407_101995688 | 0.20 |
Gm36026 |
predicted gene, 36026 |
8665 |
0.11 |
| chr11_120045840_120045991 | 0.20 |
Aatk |
apoptosis-associated tyrosine kinase |
1155 |
0.3 |
| chr6_144205160_144205647 | 0.20 |
Sox5 |
SRY (sex determining region Y)-box 5 |
976 |
0.71 |
| chr16_81200177_81200598 | 0.20 |
Ncam2 |
neural cell adhesion molecule 2 |
310 |
0.93 |
| chr9_123351055_123351227 | 0.20 |
Lars2 |
leucyl-tRNA synthetase, mitochondrial |
15786 |
0.16 |
| chr10_14759598_14760682 | 0.20 |
Nmbr |
neuromedin B receptor |
17 |
0.91 |
| chr8_12915306_12916734 | 0.20 |
Mcf2l |
mcf.2 transforming sequence-like |
45 |
0.76 |
| chr6_30957331_30957937 | 0.19 |
Klf14 |
Kruppel-like factor 14 |
1444 |
0.38 |
| chr10_23345710_23346907 | 0.19 |
Eya4 |
EYA transcriptional coactivator and phosphatase 4 |
3579 |
0.36 |
| chr19_55746711_55749175 | 0.19 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
5098 |
0.32 |
| chr15_73839069_73840200 | 0.19 |
Mroh5 |
maestro heat-like repeat family member 5 |
37 |
0.97 |
| chr11_87759834_87761999 | 0.19 |
Tspoap1 |
TSPO associated protein 1 |
329 |
0.75 |
| chr9_52141326_52141501 | 0.19 |
Mir6244 |
microRNA 6244 |
25790 |
0.16 |
| chr15_89840994_89842160 | 0.19 |
Syt10 |
synaptotagmin X |
283 |
0.93 |
| chr7_45802610_45802761 | 0.19 |
Cyth2 |
cytohesin 2 |
8203 |
0.07 |
| chr1_77507828_77508329 | 0.19 |
Epha4 |
Eph receptor A4 |
7001 |
0.19 |
| chr17_46976854_46977005 | 0.19 |
Ubr2 |
ubiquitin protein ligase E3 component n-recognin 2 |
24774 |
0.15 |
| chr19_12488341_12489388 | 0.19 |
Dtx4 |
deltex 4, E3 ubiquitin ligase |
12590 |
0.1 |
| chr9_111352762_111353061 | 0.19 |
Trank1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
21909 |
0.18 |
| chr1_151685824_151686091 | 0.19 |
Fam129a |
family with sequence similarity 129, member A |
8790 |
0.22 |
| chr10_41715763_41715983 | 0.19 |
Ccdc162 |
coiled-coil domain containing 162 |
761 |
0.57 |
| chr2_73572450_73573344 | 0.19 |
Chrna1 |
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
7441 |
0.15 |
| chr4_54945891_54946424 | 0.19 |
Zfp462 |
zinc finger protein 462 |
1109 |
0.63 |
| chr2_4152602_4153854 | 0.19 |
Frmd4a |
FERM domain containing 4A |
356 |
0.79 |
| chr2_164970287_164971705 | 0.19 |
Slc12a5 |
solute carrier family 12, member 5 |
2715 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.3 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.1 | 0.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.4 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.3 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.2 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 0.2 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.1 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.4 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0061356 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.0 | 0.1 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.2 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.4 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.0 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.3 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.0 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.0 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.1 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0072174 | metanephric tubule formation(GO:0072174) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.0 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.0 | 0.0 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.0 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.0 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.0 | GO:1904938 | dopaminergic neuron axon guidance(GO:0036514) planar cell polarity pathway involved in axon guidance(GO:1904938) |
| 0.0 | 0.0 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.0 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.0 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.0 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.0 | GO:0010911 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:1902913 | positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.0 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.0 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.0 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.6 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 3.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |